Filip Braet
ScSAM: Debiasing Morphology and Distributional Variability in Subcellular Semantic Segmentation
Jul 23, 2025Abstract:The significant morphological and distributional variability among subcellular components poses a long-standing challenge for learning-based organelle segmentation models, significantly increasing the risk of biased feature learning. Existing methods often rely on single mapping relationships, overlooking feature diversity and thereby inducing biased training. Although the Segment Anything Model (SAM) provides rich feature representations, its application to subcellular scenarios is hindered by two key challenges: (1) The variability in subcellular morphology and distribution creates gaps in the label space, leading the model to learn spurious or biased features. (2) SAM focuses on global contextual understanding and often ignores fine-grained spatial details, making it challenging to capture subtle structural alterations and cope with skewed data distributions. To address these challenges, we introduce ScSAM, a method that enhances feature robustness by fusing pre-trained SAM with Masked Autoencoder (MAE)-guided cellular prior knowledge to alleviate training bias from data imbalance. Specifically, we design a feature alignment and fusion module to align pre-trained embeddings to the same feature space and efficiently combine different representations. Moreover, we present a cosine similarity matrix-based class prompt encoder to activate class-specific features to recognize subcellular categories. Extensive experiments on diverse subcellular image datasets demonstrate that ScSAM outperforms state-of-the-art methods.
Revisiting Adaptive Cellular Recognition Under Domain Shifts: A Contextual Correspondence View
Jul 19, 2024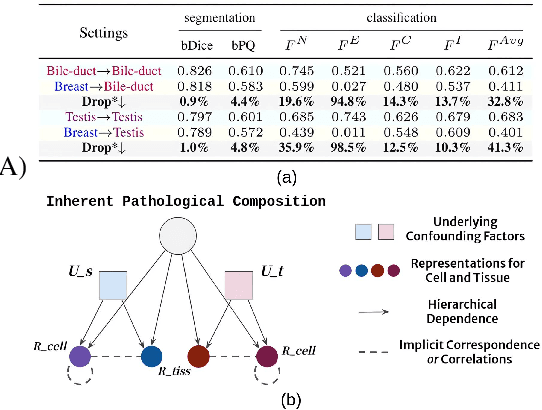
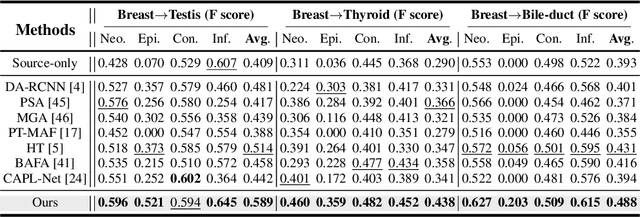
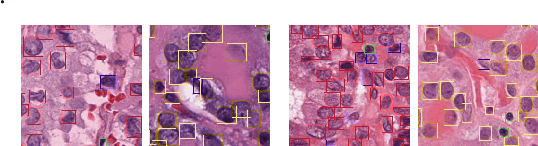
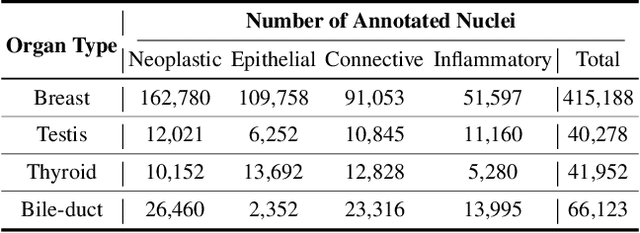
Abstract:Cellular nuclei recognition serves as a fundamental and essential step in the workflow of digital pathology. However, with disparate source organs and staining procedures among histology image clusters, the scanned tiles inherently conform to a non-uniform data distribution, which induces deteriorated promises for general cross-cohort usages. Despite the latest efforts leveraging domain adaptation to mitigate distributional discrepancy, those methods are subjected to modeling the morphological characteristics of each cell individually, disregarding the hierarchical latent structure and intrinsic contextual correspondences across the tumor micro-environment. In this work, we identify the importance of implicit correspondences across biological contexts for exploiting domain-invariant pathological composition and thereby propose to exploit the dependence over various biological structures for domain adaptive cellular recognition. We discover those high-level correspondences via unsupervised contextual modeling and use them as bridges to facilitate adaptation over diverse organs and stains. In addition, to further exploit the rich spatial contexts embedded amongst nuclear communities, we propose self-adaptive dynamic distillation to secure instance-aware trade-offs across different model constituents. The proposed method is extensively evaluated on a broad spectrum of cross-domain settings under miscellaneous data distribution shifts and outperforms the state-of-the-art methods by a substantial margin. Code is available at https://github.com/camwew/CellularRecognition_DA_CC.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge