Jianan Fan
SurgiATM: A Physics-Guided Plug-and-Play Model for Deep Learning-Based Smoke Removal in Laparoscopic Surgery
Nov 07, 2025Abstract:During laparoscopic surgery, smoke generated by tissue cauterization can significantly degrade the visual quality of endoscopic frames, increasing the risk of surgical errors and hindering both clinical decision-making and computer-assisted visual analysis. Consequently, removing surgical smoke is critical to ensuring patient safety and maintaining operative efficiency. In this study, we propose the Surgical Atmospheric Model (SurgiATM) for surgical smoke removal. SurgiATM statistically bridges a physics-based atmospheric model and data-driven deep learning models, combining the superior generalizability of the former with the high accuracy of the latter. Furthermore, SurgiATM is designed as a lightweight, plug-and-play module that can be seamlessly integrated into diverse surgical desmoking architectures to enhance their accuracy and stability, better meeting clinical requirements. It introduces only two hyperparameters and no additional trainable weights, preserving the original network architecture with minimal computational and modification overhead. We conduct extensive experiments on three public surgical datasets with ten desmoking methods, involving multiple network architectures and covering diverse procedures, including cholecystectomy, partial nephrectomy, and diaphragm dissection. The results demonstrate that incorporating SurgiATM commonly reduces the restoration errors of existing models and relatively enhances their generalizability, without adding any trainable layers or weights. This highlights the convenience, low cost, effectiveness, and generalizability of the proposed method. The code for SurgiATM is released at https://github.com/MingyuShengSMY/SurgiATM.
ScSAM: Debiasing Morphology and Distributional Variability in Subcellular Semantic Segmentation
Jul 23, 2025Abstract:The significant morphological and distributional variability among subcellular components poses a long-standing challenge for learning-based organelle segmentation models, significantly increasing the risk of biased feature learning. Existing methods often rely on single mapping relationships, overlooking feature diversity and thereby inducing biased training. Although the Segment Anything Model (SAM) provides rich feature representations, its application to subcellular scenarios is hindered by two key challenges: (1) The variability in subcellular morphology and distribution creates gaps in the label space, leading the model to learn spurious or biased features. (2) SAM focuses on global contextual understanding and often ignores fine-grained spatial details, making it challenging to capture subtle structural alterations and cope with skewed data distributions. To address these challenges, we introduce ScSAM, a method that enhances feature robustness by fusing pre-trained SAM with Masked Autoencoder (MAE)-guided cellular prior knowledge to alleviate training bias from data imbalance. Specifically, we design a feature alignment and fusion module to align pre-trained embeddings to the same feature space and efficiently combine different representations. Moreover, we present a cosine similarity matrix-based class prompt encoder to activate class-specific features to recognize subcellular categories. Extensive experiments on diverse subcellular image datasets demonstrate that ScSAM outperforms state-of-the-art methods.
Sequential Spatial-Temporal Network for Interpretable Automatic Ultrasonic Assessment of Fetal Head during labor
Mar 20, 2025Abstract:The intrapartum ultrasound guideline established by ISUOG highlights the Angle of Progression (AoP) and Head Symphysis Distance (HSD) as pivotal metrics for assessing fetal head descent and predicting delivery outcomes. Accurate measurement of the AoP and HSD requires a structured process. This begins with identifying standardized ultrasound planes, followed by the detection of specific anatomical landmarks within the regions of the pubic symphysis and fetal head that correlate with the delivery parameters AoP and HSD. Finally, these measurements are derived based on the identified anatomical landmarks. Addressing the clinical demands and standard operation process outlined in the ISUOG guideline, we introduce the Sequential Spatial-Temporal Network (SSTN), the first interpretable model specifically designed for the video of intrapartum ultrasound analysis. The SSTN operates by first identifying ultrasound planes, then segmenting anatomical structures such as the pubic symphysis and fetal head, and finally detecting key landmarks for precise measurement of HSD and AoP. Furthermore, the cohesive framework leverages task-related information to improve accuracy and reliability. Experimental evaluations on clinical datasets demonstrate that SSTN significantly surpasses existing models, reducing the mean absolute error by 18% for AoP and 22% for HSD.
Medical Image Registration Meets Vision Foundation Model: Prototype Learning and Contour Awareness
Feb 17, 2025Abstract:Medical image registration is a fundamental task in medical image analysis, aiming to establish spatial correspondences between paired images. However, existing unsupervised deformable registration methods rely solely on intensity-based similarity metrics, lacking explicit anatomical knowledge, which limits their accuracy and robustness. Vision foundation models, such as the Segment Anything Model (SAM), can generate high-quality segmentation masks that provide explicit anatomical structure knowledge, addressing the limitations of traditional methods that depend only on intensity similarity. Based on this, we propose a novel SAM-assisted registration framework incorporating prototype learning and contour awareness. The framework includes: (1) Explicit anatomical information injection, where SAM-generated segmentation masks are used as auxiliary inputs throughout training and testing to ensure the consistency of anatomical information; (2) Prototype learning, which leverages segmentation masks to extract prototype features and aligns prototypes to optimize semantic correspondences between images; and (3) Contour-aware loss, a contour-aware loss is designed that leverages the edges of segmentation masks to improve the model's performance in fine-grained deformation fields. Extensive experiments demonstrate that the proposed framework significantly outperforms existing methods across multiple datasets, particularly in challenging scenarios with complex anatomical structures and ambiguous boundaries. Our code is available at https://github.com/HaoXu0507/IPMI25-SAM-Assisted-Registration.
Cell as Point: One-Stage Framework for Efficient Cell Tracking
Nov 22, 2024



Abstract:Cellular activities are dynamic and intricate, playing a crucial role in advancing diagnostic and therapeutic techniques, yet they often require substantial resources for accurate tracking. Despite recent progress, the conventional multi-stage cell tracking approaches not only heavily rely on detection or segmentation results as a prerequisite for the tracking stage, demanding plenty of refined segmentation masks, but are also deteriorated by imbalanced and long sequence data, leading to under-learning in training and missing cells in inference procedures. To alleviate the above issues, this paper proposes the novel end-to-end CAP framework, which leverages the idea of regarding Cell as Point to achieve efficient and stable cell tracking in one stage. CAP abandons detection or segmentation stages and simplifies the process by exploiting the correlation among the trajectories of cell points to track cells jointly, thus reducing the label demand and complexity of the pipeline. With cell point trajectory and visibility to represent cell locations and lineage relationships, CAP leverages the key innovations of adaptive event-guided (AEG) sampling for addressing data imbalance in cell division events and the rolling-as-window (RAW) inference method to ensure continuous tracking of new cells in the long term. Eliminating the need for a prerequisite detection or segmentation stage, CAP demonstrates strong cell tracking performance while also being 10 to 55 times more efficient than existing methods. The code and models will be released.
AMNCutter: Affinity-Attention-Guided Multi-View Normalized Cutter for Unsupervised Surgical Instrument Segmentation
Nov 07, 2024



Abstract:Surgical instrument segmentation (SIS) is pivotal for robotic-assisted minimally invasive surgery, assisting surgeons by identifying surgical instruments in endoscopic video frames. Recent unsupervised surgical instrument segmentation (USIS) methods primarily rely on pseudo-labels derived from low-level features such as color and optical flow, but these methods show limited effectiveness and generalizability in complex and unseen endoscopic scenarios. In this work, we propose a label-free unsupervised model featuring a novel module named Multi-View Normalized Cutter (m-NCutter). Different from previous USIS works, our model is trained using a graph-cutting loss function that leverages patch affinities for supervision, eliminating the need for pseudo-labels. The framework adaptively determines which affinities from which levels should be prioritized. Therefore, the low- and high-level features and their affinities are effectively integrated to train a label-free unsupervised model, showing superior effectiveness and generalization ability. We conduct comprehensive experiments across multiple SIS datasets to validate our approach's state-of-the-art (SOTA) performance, robustness, and exceptional potential as a pre-trained model. Our code is released at https://github.com/MingyuShengSMY/AMNCutter.
Revisiting Surgical Instrument Segmentation Without Human Intervention: A Graph Partitioning View
Aug 27, 2024



Abstract:Surgical instrument segmentation (SIS) on endoscopic images stands as a long-standing and essential task in the context of computer-assisted interventions for boosting minimally invasive surgery. Given the recent surge of deep learning methodologies and their data-hungry nature, training a neural predictive model based on massive expert-curated annotations has been dominating and served as an off-the-shelf approach in the field, which could, however, impose prohibitive burden to clinicians for preparing fine-grained pixel-wise labels corresponding to the collected surgical video frames. In this work, we propose an unsupervised method by reframing the video frame segmentation as a graph partitioning problem and regarding image pixels as graph nodes, which is significantly different from the previous efforts. A self-supervised pre-trained model is firstly leveraged as a feature extractor to capture high-level semantic features. Then, Laplacian matrixs are computed from the features and are eigendecomposed for graph partitioning. On the "deep" eigenvectors, a surgical video frame is meaningfully segmented into different modules such as tools and tissues, providing distinguishable semantic information like locations, classes, and relations. The segmentation problem can then be naturally tackled by applying clustering or threshold on the eigenvectors. Extensive experiments are conducted on various datasets (e.g., EndoVis2017, EndoVis2018, UCL, etc.) for different clinical endpoints. Across all the challenging scenarios, our method demonstrates outstanding performance and robustness higher than unsupervised state-of-the-art (SOTA) methods. The code is released at https://github.com/MingyuShengSMY/GraphClusteringSIS.git.
Revisiting Adaptive Cellular Recognition Under Domain Shifts: A Contextual Correspondence View
Jul 19, 2024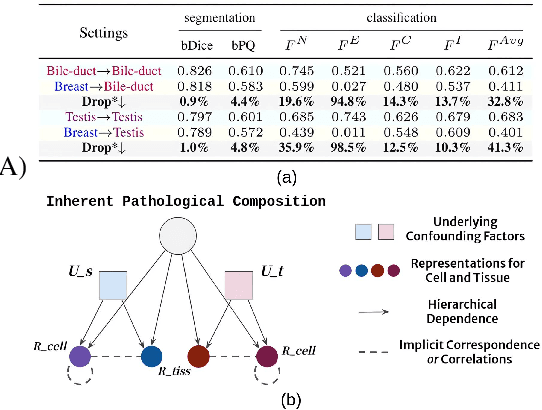
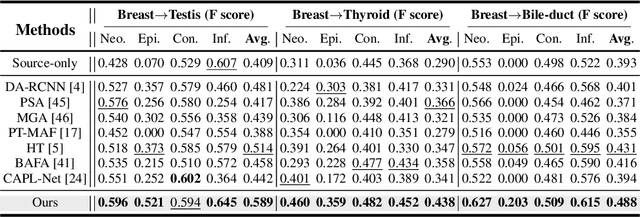
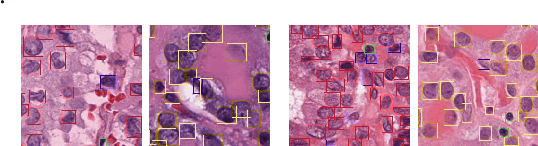
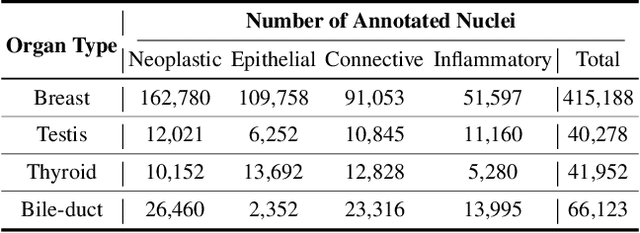
Abstract:Cellular nuclei recognition serves as a fundamental and essential step in the workflow of digital pathology. However, with disparate source organs and staining procedures among histology image clusters, the scanned tiles inherently conform to a non-uniform data distribution, which induces deteriorated promises for general cross-cohort usages. Despite the latest efforts leveraging domain adaptation to mitigate distributional discrepancy, those methods are subjected to modeling the morphological characteristics of each cell individually, disregarding the hierarchical latent structure and intrinsic contextual correspondences across the tumor micro-environment. In this work, we identify the importance of implicit correspondences across biological contexts for exploiting domain-invariant pathological composition and thereby propose to exploit the dependence over various biological structures for domain adaptive cellular recognition. We discover those high-level correspondences via unsupervised contextual modeling and use them as bridges to facilitate adaptation over diverse organs and stains. In addition, to further exploit the rich spatial contexts embedded amongst nuclear communities, we propose self-adaptive dynamic distillation to secure instance-aware trade-offs across different model constituents. The proposed method is extensively evaluated on a broad spectrum of cross-domain settings under miscellaneous data distribution shifts and outperforms the state-of-the-art methods by a substantial margin. Code is available at https://github.com/camwew/CellularRecognition_DA_CC.
Seeing Unseen: Discover Novel Biomedical Concepts via Geometry-Constrained Probabilistic Modeling
Mar 05, 2024



Abstract:Machine learning holds tremendous promise for transforming the fundamental practice of scientific discovery by virtue of its data-driven nature. With the ever-increasing stream of research data collection, it would be appealing to autonomously explore patterns and insights from observational data for discovering novel classes of phenotypes and concepts. However, in the biomedical domain, there are several challenges inherently presented in the cumulated data which hamper the progress of novel class discovery. The non-i.i.d. data distribution accompanied by the severe imbalance among different groups of classes essentially leads to ambiguous and biased semantic representations. In this work, we present a geometry-constrained probabilistic modeling treatment to resolve the identified issues. First, we propose to parameterize the approximated posterior of instance embedding as a marginal von MisesFisher distribution to account for the interference of distributional latent bias. Then, we incorporate a suite of critical geometric properties to impose proper constraints on the layout of constructed embedding space, which in turn minimizes the uncontrollable risk for unknown class learning and structuring. Furthermore, a spectral graph-theoretic method is devised to estimate the number of potential novel classes. It inherits two intriguing merits compared to existent approaches, namely high computational efficiency and flexibility for taxonomy-adaptive estimation. Extensive experiments across various biomedical scenarios substantiate the effectiveness and general applicability of our method.
Multi-source-free Domain Adaptation via Uncertainty-aware Adaptive Distillation
Feb 09, 2024

Abstract:Source-free domain adaptation (SFDA) alleviates the domain discrepancy among data obtained from domains without accessing the data for the awareness of data privacy. However, existing conventional SFDA methods face inherent limitations in medical contexts, where medical data are typically collected from multiple institutions using various equipment. To address this problem, we propose a simple yet effective method, named Uncertainty-aware Adaptive Distillation (UAD) for the multi-source-free unsupervised domain adaptation (MSFDA) setting. UAD aims to perform well-calibrated knowledge distillation from (i) model level to deliver coordinated and reliable base model initialisation and (ii) instance level via model adaptation guided by high-quality pseudo-labels, thereby obtaining a high-performance target domain model. To verify its general applicability, we evaluate UAD on two image-based diagnosis benchmarks among two multi-centre datasets, where our method shows a significant performance gain compared with existing works. The code will be available soon.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge