Ron Kikinis
MHub.ai: A Simple, Standardized, and Reproducible Platform for AI Models in Medical Imaging
Jan 15, 2026Abstract:Artificial intelligence (AI) has the potential to transform medical imaging by automating image analysis and accelerating clinical research. However, research and clinical use are limited by the wide variety of AI implementations and architectures, inconsistent documentation, and reproducibility issues. Here, we introduce MHub.ai, an open-source, container-based platform that standardizes access to AI models with minimal configuration, promoting accessibility and reproducibility in medical imaging. MHub.ai packages models from peer-reviewed publications into standardized containers that support direct processing of DICOM and other formats, provide a unified application interface, and embed structured metadata. Each model is accompanied by publicly available reference data that can be used to confirm model operation. MHub.ai includes an initial set of state-of-the-art segmentation, prediction, and feature extraction models for different modalities. The modular framework enables adaptation of any model and supports community contributions. We demonstrate the utility of the platform in a clinical use case through comparative evaluation of lung segmentation models. To further strengthen transparency and reproducibility, we publicly release the generated segmentations and evaluation metrics and provide interactive dashboards that allow readers to inspect individual cases and reproduce or extend our analysis. By simplifying model use, MHub.ai enables side-by-side benchmarking with identical execution commands and standardized outputs, and lowers the barrier to clinical translation.
In search of truth: Evaluating concordance of AI-based anatomy segmentation models
Dec 17, 2025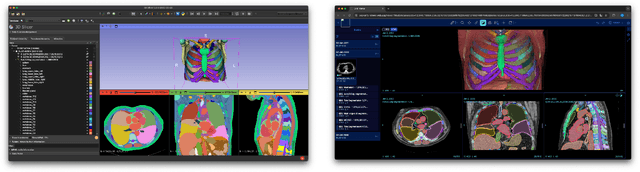


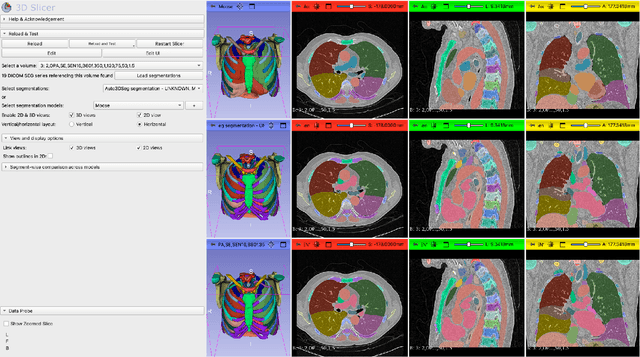
Abstract:Purpose AI-based methods for anatomy segmentation can help automate characterization of large imaging datasets. The growing number of similar in functionality models raises the challenge of evaluating them on datasets that do not contain ground truth annotations. We introduce a practical framework to assist in this task. Approach We harmonize the segmentation results into a standard, interoperable representation, which enables consistent, terminology-based labeling of the structures. We extend 3D Slicer to streamline loading and comparison of these harmonized segmentations, and demonstrate how standard representation simplifies review of the results using interactive summary plots and browser-based visualization using OHIF Viewer. To demonstrate the utility of the approach we apply it to evaluating segmentation of 31 anatomical structures (lungs, vertebrae, ribs, and heart) by six open-source models - TotalSegmentator 1.5 and 2.6, Auto3DSeg, MOOSE, MultiTalent, and CADS - for a sample of Computed Tomography (CT) scans from the publicly available National Lung Screening Trial (NLST) dataset. Results We demonstrate the utility of the framework in enabling automating loading, structure-wise inspection and comparison across models. Preliminary results ascertain practical utility of the approach in allowing quick detection and review of problematic results. The comparison shows excellent agreement segmenting some (e.g., lung) but not all structures (e.g., some models produce invalid vertebrae or rib segmentations). Conclusions The resources developed are linked from https://imagingdatacommons.github.io/segmentation-comparison/ including segmentation harmonization scripts, summary plots, and visualization tools. This work assists in model evaluation in absence of ground truth, ultimately enabling informed model selection.
SurgiATM: A Physics-Guided Plug-and-Play Model for Deep Learning-Based Smoke Removal in Laparoscopic Surgery
Nov 07, 2025Abstract:During laparoscopic surgery, smoke generated by tissue cauterization can significantly degrade the visual quality of endoscopic frames, increasing the risk of surgical errors and hindering both clinical decision-making and computer-assisted visual analysis. Consequently, removing surgical smoke is critical to ensuring patient safety and maintaining operative efficiency. In this study, we propose the Surgical Atmospheric Model (SurgiATM) for surgical smoke removal. SurgiATM statistically bridges a physics-based atmospheric model and data-driven deep learning models, combining the superior generalizability of the former with the high accuracy of the latter. Furthermore, SurgiATM is designed as a lightweight, plug-and-play module that can be seamlessly integrated into diverse surgical desmoking architectures to enhance their accuracy and stability, better meeting clinical requirements. It introduces only two hyperparameters and no additional trainable weights, preserving the original network architecture with minimal computational and modification overhead. We conduct extensive experiments on three public surgical datasets with ten desmoking methods, involving multiple network architectures and covering diverse procedures, including cholecystectomy, partial nephrectomy, and diaphragm dissection. The results demonstrate that incorporating SurgiATM commonly reduces the restoration errors of existing models and relatively enhances their generalizability, without adding any trainable layers or weights. This highlights the convenience, low cost, effectiveness, and generalizability of the proposed method. The code for SurgiATM is released at https://github.com/MingyuShengSMY/SurgiATM.
Benchmarking of Deep Learning Methods for Generic MRI Multi-OrganAbdominal Segmentation
Jul 23, 2025Abstract:Recent advances in deep learning have led to robust automated tools for segmentation of abdominal computed tomography (CT). Meanwhile, segmentation of magnetic resonance imaging (MRI) is substantially more challenging due to the inherent signal variability and the increased effort required for annotating training datasets. Hence, existing approaches are trained on limited sets of MRI sequences, which might limit their generalizability. To characterize the landscape of MRI abdominal segmentation tools, we present here a comprehensive benchmarking of the three state-of-the-art and open-source models: MRSegmentator, MRISegmentator-Abdomen, and TotalSegmentator MRI. Since these models are trained using labor-intensive manual annotation cycles, we also introduce and evaluate ABDSynth, a SynthSeg-based model purely trained on widely available CT segmentations (no real images). More generally, we assess accuracy and generalizability by leveraging three public datasets (not seen by any of the evaluated methods during their training), which span all major manufacturers, five MRI sequences, as well as a variety of subject conditions, voxel resolutions, and fields-of-view. Our results reveal that MRSegmentator achieves the best performance and is most generalizable. In contrast, ABDSynth yields slightly less accurate results, but its relaxed requirements in training data make it an alternative when the annotation budget is limited. The evaluation code and datasets are given for future benchmarking at https://github.com/deepakri201/AbdoBench, along with inference code and weights for ABDSynth.
MultiCo3D: Multi-Label Voxel Contrast for One-Shot Incremental Segmentation of 3D Neuroimages
Mar 09, 2025Abstract:3D neuroimages provide a comprehensive view of brain structure and function, aiding in precise localization and functional connectivity analysis. Segmentation of white matter (WM) tracts using 3D neuroimages is vital for understanding the brain's structural connectivity in both healthy and diseased states. One-shot Class Incremental Semantic Segmentation (OCIS) refers to effectively segmenting new (novel) classes using only a single sample while retaining knowledge of old (base) classes without forgetting. Voxel-contrastive OCIS methods adjust the feature space to alleviate the feature overlap problem between the base and novel classes. However, since WM tract segmentation is a multi-label segmentation task, existing single-label voxel contrastive-based methods may cause inherent contradictions. To address this, we propose a new multi-label voxel contrast framework called MultiCo3D for one-shot class incremental tract segmentation. Our method utilizes uncertainty distillation to preserve base tract segmentation knowledge while adjusting the feature space with multi-label voxel contrast to alleviate feature overlap when learning novel tracts and dynamically weighting multi losses to balance overall loss. We compare our method against several state-of-the-art (SOTA) approaches. The experimental results show that our method significantly enhances one-shot class incremental tract segmentation accuracy across five different experimental setups on HCP and Preto datasets.
Medical Image Registration Meets Vision Foundation Model: Prototype Learning and Contour Awareness
Feb 17, 2025Abstract:Medical image registration is a fundamental task in medical image analysis, aiming to establish spatial correspondences between paired images. However, existing unsupervised deformable registration methods rely solely on intensity-based similarity metrics, lacking explicit anatomical knowledge, which limits their accuracy and robustness. Vision foundation models, such as the Segment Anything Model (SAM), can generate high-quality segmentation masks that provide explicit anatomical structure knowledge, addressing the limitations of traditional methods that depend only on intensity similarity. Based on this, we propose a novel SAM-assisted registration framework incorporating prototype learning and contour awareness. The framework includes: (1) Explicit anatomical information injection, where SAM-generated segmentation masks are used as auxiliary inputs throughout training and testing to ensure the consistency of anatomical information; (2) Prototype learning, which leverages segmentation masks to extract prototype features and aligns prototypes to optimize semantic correspondences between images; and (3) Contour-aware loss, a contour-aware loss is designed that leverages the edges of segmentation masks to improve the model's performance in fine-grained deformation fields. Extensive experiments demonstrate that the proposed framework significantly outperforms existing methods across multiple datasets, particularly in challenging scenarios with complex anatomical structures and ambiguous boundaries. Our code is available at https://github.com/HaoXu0507/IPMI25-SAM-Assisted-Registration.
Tab2Visual: Overcoming Limited Data in Tabular Data Classification Using Deep Learning with Visual Representations
Feb 11, 2025Abstract:This research addresses the challenge of limited data in tabular data classification, particularly prevalent in domains with constraints like healthcare. We propose Tab2Visual, a novel approach that transforms heterogeneous tabular data into visual representations, enabling the application of powerful deep learning models. Tab2Visual effectively addresses data scarcity by incorporating novel image augmentation techniques and facilitating transfer learning. We extensively evaluate the proposed approach on diverse tabular datasets, comparing its performance against a wide range of machine learning algorithms, including classical methods, tree-based ensembles, and state-of-the-art deep learning models specifically designed for tabular data. We also perform an in-depth analysis of factors influencing Tab2Visual's performance. Our experimental results demonstrate that Tab2Visual outperforms other methods in classification problems with limited tabular data.
AMNCutter: Affinity-Attention-Guided Multi-View Normalized Cutter for Unsupervised Surgical Instrument Segmentation
Nov 07, 2024



Abstract:Surgical instrument segmentation (SIS) is pivotal for robotic-assisted minimally invasive surgery, assisting surgeons by identifying surgical instruments in endoscopic video frames. Recent unsupervised surgical instrument segmentation (USIS) methods primarily rely on pseudo-labels derived from low-level features such as color and optical flow, but these methods show limited effectiveness and generalizability in complex and unseen endoscopic scenarios. In this work, we propose a label-free unsupervised model featuring a novel module named Multi-View Normalized Cutter (m-NCutter). Different from previous USIS works, our model is trained using a graph-cutting loss function that leverages patch affinities for supervision, eliminating the need for pseudo-labels. The framework adaptively determines which affinities from which levels should be prioritized. Therefore, the low- and high-level features and their affinities are effectively integrated to train a label-free unsupervised model, showing superior effectiveness and generalization ability. We conduct comprehensive experiments across multiple SIS datasets to validate our approach's state-of-the-art (SOTA) performance, robustness, and exceptional potential as a pre-trained model. Our code is released at https://github.com/MingyuShengSMY/AMNCutter.
Revisiting Surgical Instrument Segmentation Without Human Intervention: A Graph Partitioning View
Aug 27, 2024



Abstract:Surgical instrument segmentation (SIS) on endoscopic images stands as a long-standing and essential task in the context of computer-assisted interventions for boosting minimally invasive surgery. Given the recent surge of deep learning methodologies and their data-hungry nature, training a neural predictive model based on massive expert-curated annotations has been dominating and served as an off-the-shelf approach in the field, which could, however, impose prohibitive burden to clinicians for preparing fine-grained pixel-wise labels corresponding to the collected surgical video frames. In this work, we propose an unsupervised method by reframing the video frame segmentation as a graph partitioning problem and regarding image pixels as graph nodes, which is significantly different from the previous efforts. A self-supervised pre-trained model is firstly leveraged as a feature extractor to capture high-level semantic features. Then, Laplacian matrixs are computed from the features and are eigendecomposed for graph partitioning. On the "deep" eigenvectors, a surgical video frame is meaningfully segmented into different modules such as tools and tissues, providing distinguishable semantic information like locations, classes, and relations. The segmentation problem can then be naturally tackled by applying clustering or threshold on the eigenvectors. Extensive experiments are conducted on various datasets (e.g., EndoVis2017, EndoVis2018, UCL, etc.) for different clinical endpoints. Across all the challenging scenarios, our method demonstrates outstanding performance and robustness higher than unsupervised state-of-the-art (SOTA) methods. The code is released at https://github.com/MingyuShengSMY/GraphClusteringSIS.git.
LNQ 2023 challenge: Benchmark of weakly-supervised techniques for mediastinal lymph node quantification
Aug 19, 2024



Abstract:Accurate assessment of lymph node size in 3D CT scans is crucial for cancer staging, therapeutic management, and monitoring treatment response. Existing state-of-the-art segmentation frameworks in medical imaging often rely on fully annotated datasets. However, for lymph node segmentation, these datasets are typically small due to the extensive time and expertise required to annotate the numerous lymph nodes in 3D CT scans. Weakly-supervised learning, which leverages incomplete or noisy annotations, has recently gained interest in the medical imaging community as a potential solution. Despite the variety of weakly-supervised techniques proposed, most have been validated only on private datasets or small publicly available datasets. To address this limitation, the Mediastinal Lymph Node Quantification (LNQ) challenge was organized in conjunction with the 26th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2023). This challenge aimed to advance weakly-supervised segmentation methods by providing a new, partially annotated dataset and a robust evaluation framework. A total of 16 teams from 5 countries submitted predictions to the validation leaderboard, and 6 teams from 3 countries participated in the evaluation phase. The results highlighted both the potential and the current limitations of weakly-supervised approaches. On one hand, weakly-supervised approaches obtained relatively good performance with a median Dice score of $61.0\%$. On the other hand, top-ranked teams, with a median Dice score exceeding $70\%$, boosted their performance by leveraging smaller but fully annotated datasets to combine weak supervision and full supervision. This highlights both the promise of weakly-supervised methods and the ongoing need for high-quality, fully annotated data to achieve higher segmentation performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge