Deepa Krishnaswamy
In search of truth: Evaluating concordance of AI-based anatomy segmentation models
Dec 17, 2025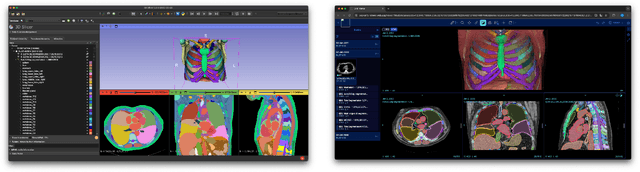


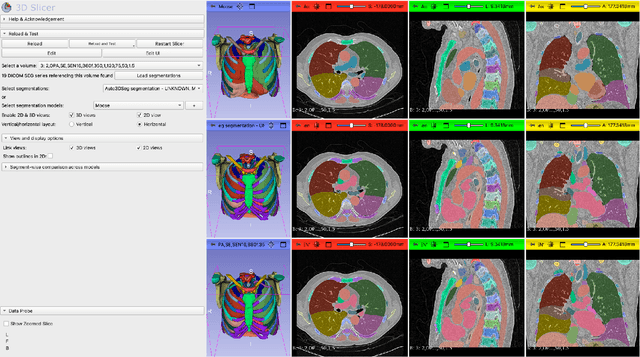
Abstract:Purpose AI-based methods for anatomy segmentation can help automate characterization of large imaging datasets. The growing number of similar in functionality models raises the challenge of evaluating them on datasets that do not contain ground truth annotations. We introduce a practical framework to assist in this task. Approach We harmonize the segmentation results into a standard, interoperable representation, which enables consistent, terminology-based labeling of the structures. We extend 3D Slicer to streamline loading and comparison of these harmonized segmentations, and demonstrate how standard representation simplifies review of the results using interactive summary plots and browser-based visualization using OHIF Viewer. To demonstrate the utility of the approach we apply it to evaluating segmentation of 31 anatomical structures (lungs, vertebrae, ribs, and heart) by six open-source models - TotalSegmentator 1.5 and 2.6, Auto3DSeg, MOOSE, MultiTalent, and CADS - for a sample of Computed Tomography (CT) scans from the publicly available National Lung Screening Trial (NLST) dataset. Results We demonstrate the utility of the framework in enabling automating loading, structure-wise inspection and comparison across models. Preliminary results ascertain practical utility of the approach in allowing quick detection and review of problematic results. The comparison shows excellent agreement segmenting some (e.g., lung) but not all structures (e.g., some models produce invalid vertebrae or rib segmentations). Conclusions The resources developed are linked from https://imagingdatacommons.github.io/segmentation-comparison/ including segmentation harmonization scripts, summary plots, and visualization tools. This work assists in model evaluation in absence of ground truth, ultimately enabling informed model selection.
Benchmarking of Deep Learning Methods for Generic MRI Multi-OrganAbdominal Segmentation
Jul 23, 2025Abstract:Recent advances in deep learning have led to robust automated tools for segmentation of abdominal computed tomography (CT). Meanwhile, segmentation of magnetic resonance imaging (MRI) is substantially more challenging due to the inherent signal variability and the increased effort required for annotating training datasets. Hence, existing approaches are trained on limited sets of MRI sequences, which might limit their generalizability. To characterize the landscape of MRI abdominal segmentation tools, we present here a comprehensive benchmarking of the three state-of-the-art and open-source models: MRSegmentator, MRISegmentator-Abdomen, and TotalSegmentator MRI. Since these models are trained using labor-intensive manual annotation cycles, we also introduce and evaluate ABDSynth, a SynthSeg-based model purely trained on widely available CT segmentations (no real images). More generally, we assess accuracy and generalizability by leveraging three public datasets (not seen by any of the evaluated methods during their training), which span all major manufacturers, five MRI sequences, as well as a variety of subject conditions, voxel resolutions, and fields-of-view. Our results reveal that MRSegmentator achieves the best performance and is most generalizable. In contrast, ABDSynth yields slightly less accurate results, but its relaxed requirements in training data make it an alternative when the annotation budget is limited. The evaluation code and datasets are given for future benchmarking at https://github.com/deepakri201/AbdoBench, along with inference code and weights for ABDSynth.
Rule-based outlier detection of AI-generated anatomy segmentations
Jun 20, 2024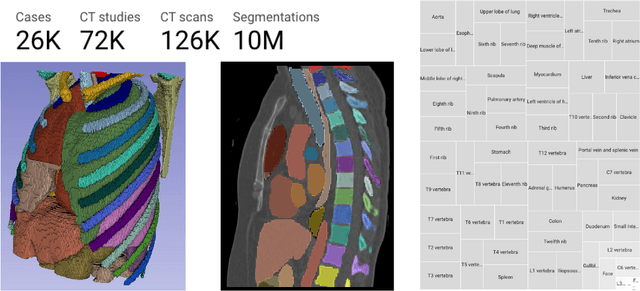
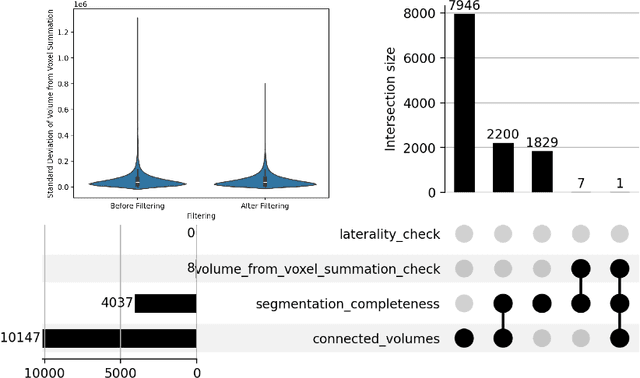
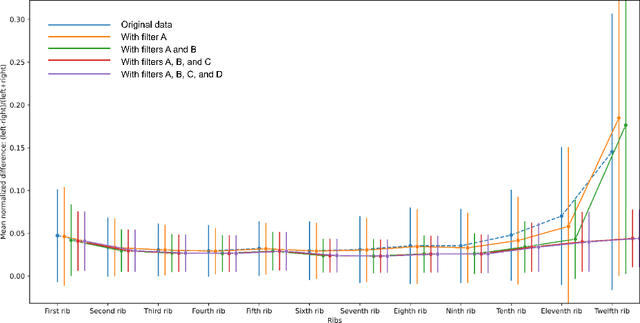
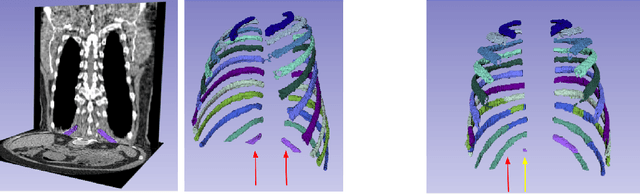
Abstract:There is a dire need for medical imaging datasets with accompanying annotations to perform downstream patient analysis. However, it is difficult to manually generate these annotations, due to the time-consuming nature, and the variability in clinical conventions. Artificial intelligence has been adopted in the field as a potential method to annotate these large datasets, however, a lack of expert annotations or ground truth can inhibit the adoption of these annotations. We recently made a dataset publicly available including annotations and extracted features of up to 104 organs for the National Lung Screening Trial using the TotalSegmentator method. However, the released dataset does not include expert-derived annotations or an assessment of the accuracy of the segmentations, limiting its usefulness. We propose the development of heuristics to assess the quality of the segmentations, providing methods to measure the consistency of the annotations and a comparison of results to the literature. We make our code and related materials publicly available at https://github.com/ImagingDataCommons/CloudSegmentatorResults and interactive tools at https://huggingface.co/spaces/ImagingDataCommons/CloudSegmentatorResults.
Automatic classification of prostate MR series type using image content and metadata
Apr 16, 2024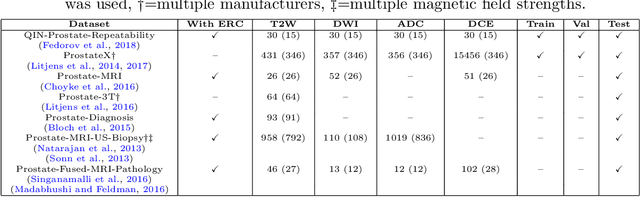
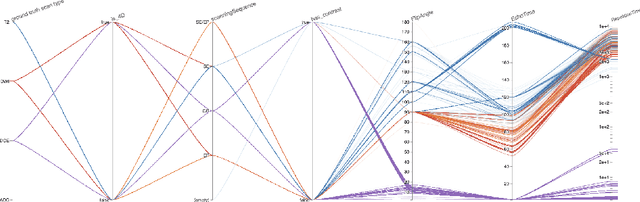
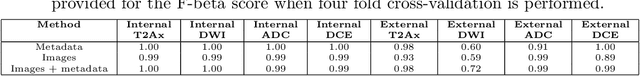
Abstract:With the wealth of medical image data, efficient curation is essential. Assigning the sequence type to magnetic resonance images is necessary for scientific studies and artificial intelligence-based analysis. However, incomplete or missing metadata prevents effective automation. We therefore propose a deep-learning method for classification of prostate cancer scanning sequences based on a combination of image data and DICOM metadata. We demonstrate superior results compared to metadata or image data alone, and make our code publicly available at https://github.com/deepakri201/DICOMScanClassification.
Towards Automatic Abdominal MRI Organ Segmentation: Leveraging Synthesized Data Generated From CT Labels
Mar 22, 2024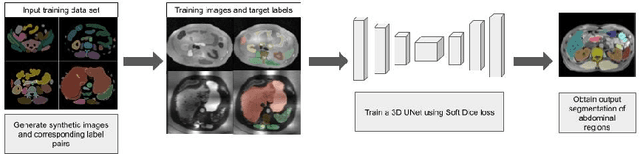

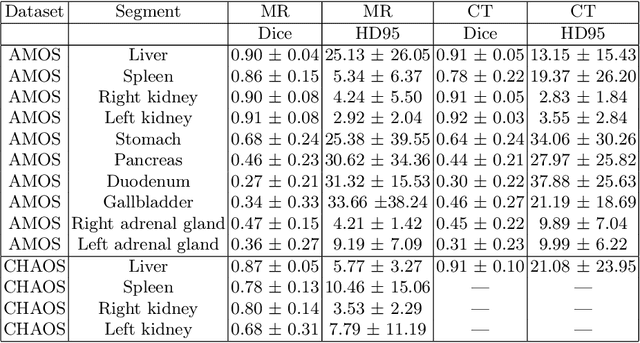
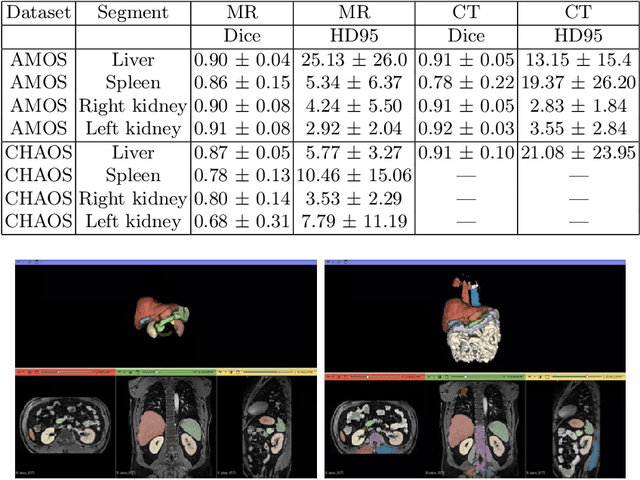
Abstract:Deep learning has shown great promise in the ability to automatically annotate organs in magnetic resonance imaging (MRI) scans, for example, of the brain. However, despite advancements in the field, the ability to accurately segment abdominal organs remains difficult across MR. In part, this may be explained by the much greater variability in image appearance and severely limited availability of training labels. The inherent nature of computed tomography (CT) scans makes it easier to annotate, resulting in a larger availability of expert annotations for the latter. We leverage a modality-agnostic domain randomization approach, utilizing CT label maps to generate synthetic images on-the-fly during training, further used to train a U-Net segmentation network for abdominal organs segmentation. Our approach shows comparable results compared to fully-supervised segmentation methods trained on MR data. Our method results in Dice scores of 0.90 (0.08) and 0.91 (0.08) for the right and left kidney respectively, compared to a pretrained nnU-Net model yielding 0.87 (0.20) and 0.91 (0.03). We will make our code publicly available.
Enrichment of the NLST and NSCLC-Radiomics computed tomography collections with AI-derived annotations
May 31, 2023Abstract:Public imaging datasets are critical for the development and evaluation of automated tools in cancer imaging. Unfortunately, many do not include annotations or image-derived features, complicating their downstream analysis. Artificial intelligence-based annotation tools have been shown to achieve acceptable performance and thus can be used to automatically annotate large datasets. As part of the effort to enrich public data available within NCI Imaging Data Commons (IDC), here we introduce AI-generated annotations for two collections of computed tomography images of the chest, NSCLC-Radiomics, and the National Lung Screening Trial. Using publicly available AI algorithms we derived volumetric annotations of thoracic organs at risk, their corresponding radiomics features, and slice-level annotations of anatomical landmarks and regions. The resulting annotations are publicly available within IDC, where the DICOM format is used to harmonize the data and achieve FAIR principles. The annotations are accompanied by cloud-enabled notebooks demonstrating their use. This study reinforces the need for large, publicly accessible curated datasets and demonstrates how AI can be used to aid in cancer imaging.
Unsupervised diffeomorphic cardiac image registration using parameterization of the deformation field
Aug 28, 2022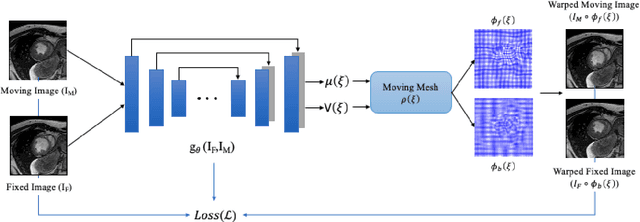

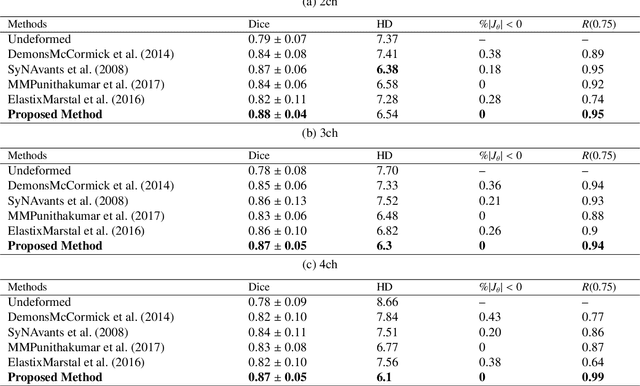

Abstract:This study proposes an end-to-end unsupervised diffeomorphic deformable registration framework based on moving mesh parameterization. Using this parameterization, a deformation field can be modeled with its transformation Jacobian determinant and curl of end velocity field. The new model of the deformation field has three important advantages; firstly, it relaxes the need for an explicit regularization term and the corresponding weight in the cost function. The smoothness is implicitly embedded in the solution which results in a physically plausible deformation field. Secondly, it guarantees diffeomorphism through explicit constraints applied to the transformation Jacobian determinant to keep it positive. Finally, it is suitable for cardiac data processing, since the nature of this parameterization is to define the deformation field in terms of the radial and rotational components. The effectiveness of the algorithm is investigated by evaluating the proposed method on three different data sets including 2D and 3D cardiac MRI scans. The results demonstrate that the proposed framework outperforms existing learning-based and non-learning-based methods while generating diffeomorphic transformations.
A New Semi-Automated Algorithm for Volumetric Segmentation of the Left Ventricle in Temporal 3D Echocardiography Sequences
Sep 03, 2021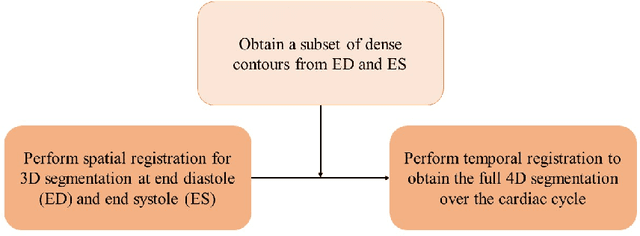
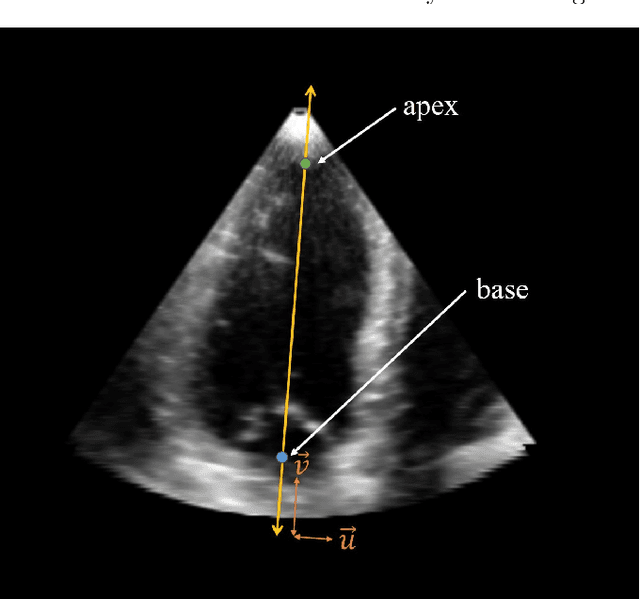
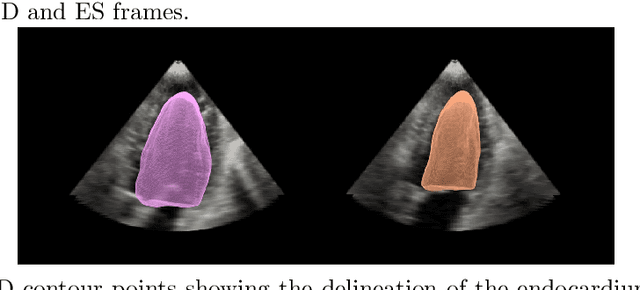
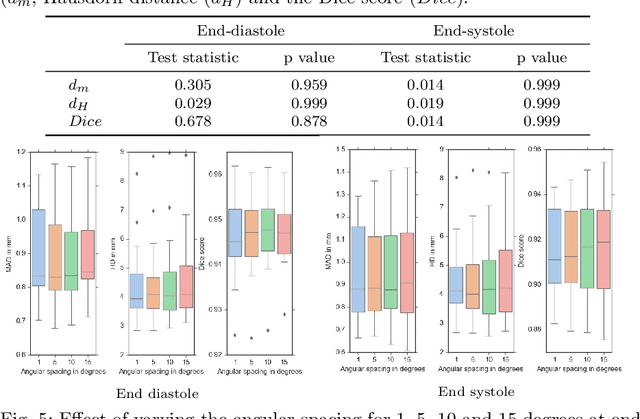
Abstract:Purpose: Echocardiography is commonly used as a non-invasive imaging tool in clinical practice for the assessment of cardiac function. However, delineation of the left ventricle is challenging due to the inherent properties of ultrasound imaging, such as the presence of speckle noise and the low signal-to-noise ratio. Methods: We propose a semi-automated segmentation algorithm for the delineation of the left ventricle in temporal 3D echocardiography sequences. The method requires minimal user interaction and relies on a diffeomorphic registration approach. Advantages of the method include no dependence on prior geometrical information, training data, or registration from an atlas. Results: The method was evaluated using three-dimensional ultrasound scan sequences from 18 patients from the Mazankowski Alberta Heart Institute, Edmonton, Canada, and compared to manual delineations provided by an expert cardiologist and four other registration algorithms. The segmentation approach yielded the following results over the cardiac cycle: a mean absolute difference of 1.01 (0.21) mm, a Hausdorff distance of 4.41 (1.43) mm, and a Dice overlap score of 0.93 (0.02). Conclusions: The method performed well compared to the four other registration algorithms.
* 22 pages, 8 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge