Hugo JWL Aerts
MHub.ai: A Simple, Standardized, and Reproducible Platform for AI Models in Medical Imaging
Jan 15, 2026Abstract:Artificial intelligence (AI) has the potential to transform medical imaging by automating image analysis and accelerating clinical research. However, research and clinical use are limited by the wide variety of AI implementations and architectures, inconsistent documentation, and reproducibility issues. Here, we introduce MHub.ai, an open-source, container-based platform that standardizes access to AI models with minimal configuration, promoting accessibility and reproducibility in medical imaging. MHub.ai packages models from peer-reviewed publications into standardized containers that support direct processing of DICOM and other formats, provide a unified application interface, and embed structured metadata. Each model is accompanied by publicly available reference data that can be used to confirm model operation. MHub.ai includes an initial set of state-of-the-art segmentation, prediction, and feature extraction models for different modalities. The modular framework enables adaptation of any model and supports community contributions. We demonstrate the utility of the platform in a clinical use case through comparative evaluation of lung segmentation models. To further strengthen transparency and reproducibility, we publicly release the generated segmentations and evaluation metrics and provide interactive dashboards that allow readers to inspect individual cases and reproduce or extend our analysis. By simplifying model use, MHub.ai enables side-by-side benchmarking with identical execution commands and standardized outputs, and lowers the barrier to clinical translation.
Foundation Artificial Intelligence Models for Health Recognition Using Face Photographs (FAHR-Face)
Jun 17, 2025Abstract:Background: Facial appearance offers a noninvasive window into health. We built FAHR-Face, a foundation model trained on >40 million facial images and fine-tuned it for two distinct tasks: biological age estimation (FAHR-FaceAge) and survival risk prediction (FAHR-FaceSurvival). Methods: FAHR-FaceAge underwent a two-stage, age-balanced fine-tuning on 749,935 public images; FAHR-FaceSurvival was fine-tuned on 34,389 photos of cancer patients. Model robustness (cosmetic surgery, makeup, pose, lighting) and independence (saliency mapping) was tested extensively. Both models were clinically tested in two independent cancer patient datasets with survival analyzed by multivariable Cox models and adjusted for clinical prognostic factors. Findings: For age estimation, FAHR-FaceAge had the lowest mean absolute error of 5.1 years on public datasets, outperforming benchmark models and maintaining accuracy across the full human lifespan. In cancer patients, FAHR-FaceAge outperformed a prior facial age estimation model in survival prognostication. FAHR-FaceSurvival demonstrated robust prediction of mortality, and the highest-risk quartile had more than triple the mortality of the lowest (adjusted hazard ratio 3.22; P<0.001). These findings were validated in the independent cohort and both models showed generalizability across age, sex, race and cancer subgroups. The two algorithms provided distinct, complementary prognostic information; saliency mapping revealed each model relied on distinct facial regions. The combination of FAHR-FaceAge and FAHR-FaceSurvival improved prognostic accuracy. Interpretation: A single foundation model can generate inexpensive, scalable facial biomarkers that capture both biological ageing and disease-related mortality risk. The foundation model enabled effective training using relatively small clinical datasets.
Artificial Intelligence-Based Opportunistic Coronary Calcium Screening in the Veterans Affairs National Healthcare System
Sep 16, 2024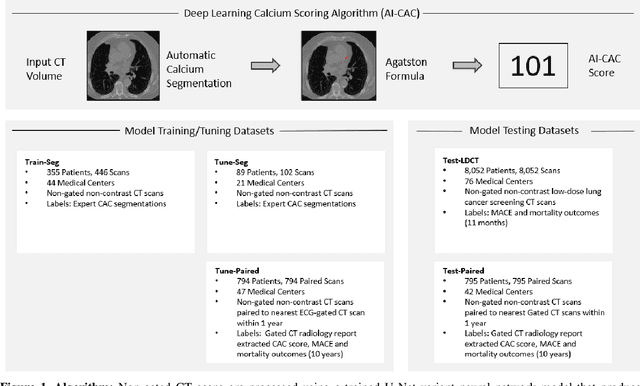
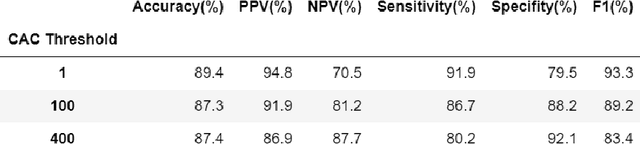
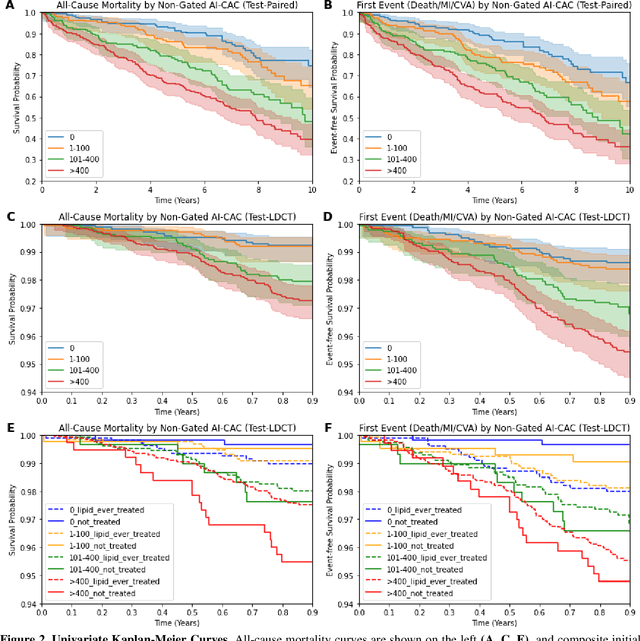
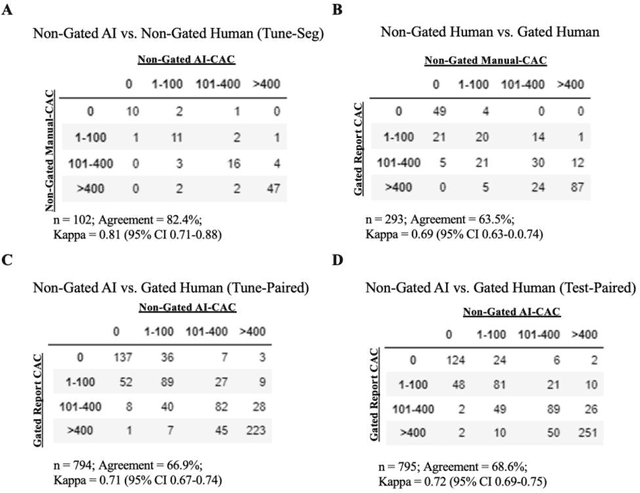
Abstract:Coronary artery calcium (CAC) is highly predictive of cardiovascular events. While millions of chest CT scans are performed annually in the United States, CAC is not routinely quantified from scans done for non-cardiac purposes. A deep learning algorithm was developed using 446 expert segmentations to automatically quantify CAC on non-contrast, non-gated CT scans (AI-CAC). Our study differs from prior works as we leverage imaging data across the Veterans Affairs national healthcare system, from 98 medical centers, capturing extensive heterogeneity in imaging protocols, scanners, and patients. AI-CAC performance on non-gated scans was compared against clinical standard ECG-gated CAC scoring. Non-gated AI-CAC differentiated zero vs. non-zero and less than 100 vs. 100 or greater Agatston scores with accuracies of 89.4% (F1 0.93) and 87.3% (F1 0.89), respectively, in 795 patients with paired gated scans within a year of a non-gated CT scan. Non-gated AI-CAC was predictive of 10-year all-cause mortality (CAC 0 vs. >400 group: 25.4% vs. 60.2%, Cox HR 3.49, p < 0.005), and composite first-time stroke, MI, or death (CAC 0 vs. >400 group: 33.5% vs. 63.8%, Cox HR 3.00, p < 0.005). In a screening dataset of 8,052 patients with low-dose lung cancer-screening CTs (LDCT), 3,091/8,052 (38.4%) individuals had AI-CAC >400. Four cardiologists qualitatively reviewed LDCT images from a random sample of >400 AI-CAC patients and verified that 527/531 (99.2%) would benefit from lipid-lowering therapy. To the best of our knowledge, this is the first non-gated CT CAC algorithm developed across a national healthcare system, on multiple imaging protocols, without filtering intra-cardiac hardware, and compared against a strong gated CT reference. We report superior performance relative to previous CAC algorithms evaluated against paired gated scans that included patients with intra-cardiac hardware.
Large Language Models to Identify Social Determinants of Health in Electronic Health Records
Aug 11, 2023Abstract:Social determinants of health (SDoH) have an important impact on patient outcomes but are incompletely collected from the electronic health records (EHR). This study researched the ability of large language models to extract SDoH from free text in EHRs, where they are most commonly documented, and explored the role of synthetic clinical text for improving the extraction of these scarcely documented, yet extremely valuable, clinical data. 800 patient notes were annotated for SDoH categories, and several transformer-based models were evaluated. The study also experimented with synthetic data generation and assessed for algorithmic bias. Our best-performing models were fine-tuned Flan-T5 XL (macro-F1 0.71) for any SDoH, and Flan-T5 XXL (macro-F1 0.70). The benefit of augmenting fine-tuning with synthetic data varied across model architecture and size, with smaller Flan-T5 models (base and large) showing the greatest improvements in performance (delta F1 +0.12 to +0.23). Model performance was similar on the in-hospital system dataset but worse on the MIMIC-III dataset. Our best-performing fine-tuned models outperformed zero- and few-shot performance of ChatGPT-family models for both tasks. These fine-tuned models were less likely than ChatGPT to change their prediction when race/ethnicity and gender descriptors were added to the text, suggesting less algorithmic bias (p<0.05). At the patient-level, our models identified 93.8% of patients with adverse SDoH, while ICD-10 codes captured 2.0%. Our method can effectively extracted SDoH information from clinic notes, performing better compare to GPT zero- and few-shot settings. These models could enhance real-world evidence on SDoH and aid in identifying patients needing social support.
Enrichment of the NLST and NSCLC-Radiomics computed tomography collections with AI-derived annotations
May 31, 2023Abstract:Public imaging datasets are critical for the development and evaluation of automated tools in cancer imaging. Unfortunately, many do not include annotations or image-derived features, complicating their downstream analysis. Artificial intelligence-based annotation tools have been shown to achieve acceptable performance and thus can be used to automatically annotate large datasets. As part of the effort to enrich public data available within NCI Imaging Data Commons (IDC), here we introduce AI-generated annotations for two collections of computed tomography images of the chest, NSCLC-Radiomics, and the National Lung Screening Trial. Using publicly available AI algorithms we derived volumetric annotations of thoracic organs at risk, their corresponding radiomics features, and slice-level annotations of anatomical landmarks and regions. The resulting annotations are publicly available within IDC, where the DICOM format is used to harmonize the data and achieve FAIR principles. The annotations are accompanied by cloud-enabled notebooks demonstrating their use. This study reinforces the need for large, publicly accessible curated datasets and demonstrates how AI can be used to aid in cancer imaging.
Evaluation of ChatGPT Family of Models for Biomedical Reasoning and Classification
Apr 05, 2023



Abstract:Recent advances in large language models (LLMs) have shown impressive ability in biomedical question-answering, but have not been adequately investigated for more specific biomedical applications. This study investigates the performance of LLMs such as the ChatGPT family of models (GPT-3.5s, GPT-4) in biomedical tasks beyond question-answering. Because no patient data can be passed to the OpenAI API public interface, we evaluated model performance with over 10000 samples as proxies for two fundamental tasks in the clinical domain - classification and reasoning. The first task is classifying whether statements of clinical and policy recommendations in scientific literature constitute health advice. The second task is causal relation detection from the biomedical literature. We compared LLMs with simpler models, such as bag-of-words (BoW) with logistic regression, and fine-tuned BioBERT models. Despite the excitement around viral ChatGPT, we found that fine-tuning for two fundamental NLP tasks remained the best strategy. The simple BoW model performed on par with the most complex LLM prompting. Prompt engineering required significant investment.
Natural language processing to automatically extract the presence and severity of esophagitis in notes of patients undergoing radiotherapy
Mar 24, 2023Abstract:Radiotherapy (RT) toxicities can impair survival and quality-of-life, yet remain under-studied. Real-world evidence holds potential to improve our understanding of toxicities, but toxicity information is often only in clinical notes. We developed natural language processing (NLP) models to identify the presence and severity of esophagitis from notes of patients treated with thoracic RT. We fine-tuned statistical and pre-trained BERT-based models for three esophagitis classification tasks: Task 1) presence of esophagitis, Task 2) severe esophagitis or not, and Task 3) no esophagitis vs. grade 1 vs. grade 2-3. Transferability was tested on 345 notes from patients with esophageal cancer undergoing RT. Fine-tuning PubmedBERT yielded the best performance. The best macro-F1 was 0.92, 0.82, and 0.74 for Task 1, 2, and 3, respectively. Selecting the most informative note sections during fine-tuning improved macro-F1 by over 2% for all tasks. Silver-labeled data improved the macro-F1 by over 3% across all tasks. For the esophageal cancer notes, the best macro-F1 was 0.73, 0.74, and 0.65 for Task 1, 2, and 3, respectively, without additional fine-tuning. To our knowledge, this is the first effort to automatically extract esophagitis toxicity severity according to CTCAE guidelines from clinic notes. The promising performance provides proof-of-concept for NLP-based automated detailed toxicity monitoring in expanded domains.
ModelHub.AI: Dissemination Platform for Deep Learning Models
Nov 26, 2019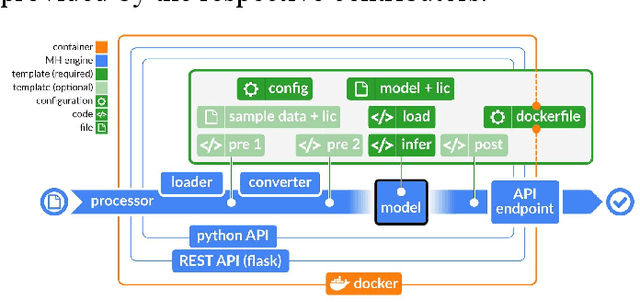
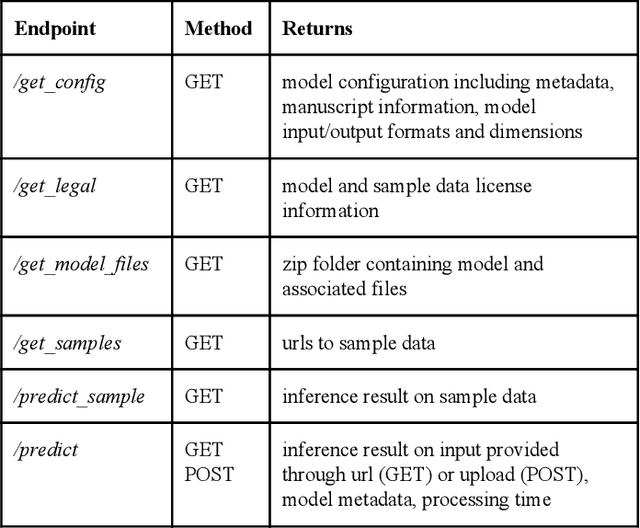
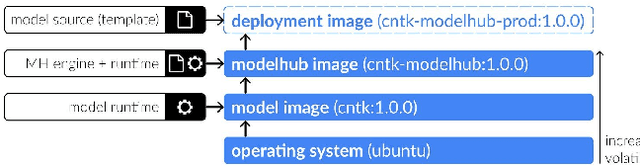
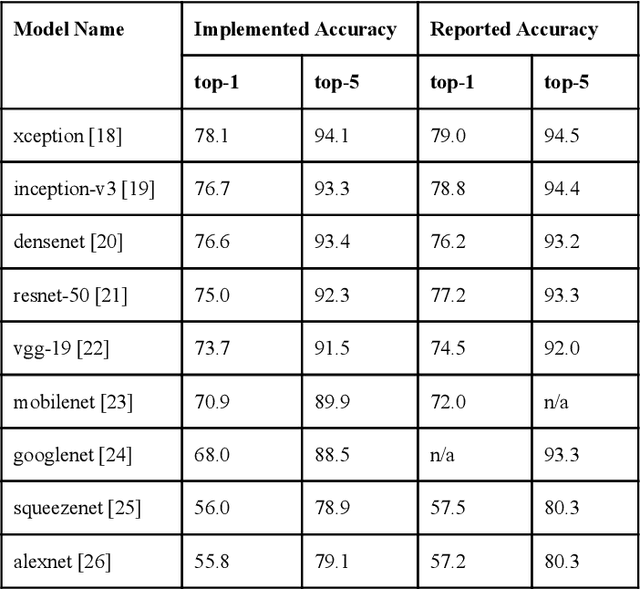
Abstract:Recent advances in artificial intelligence research have led to a profusion of studies that apply deep learning to problems in image analysis and natural language processing among others. Additionally, the availability of open-source computational frameworks has lowered the barriers to implementing state-of-the-art methods across multiple domains. Albeit leading to major performance breakthroughs in some tasks, effective dissemination of deep learning algorithms remains challenging, inhibiting reproducibility and benchmarking studies, impeding further validation, and ultimately hindering their effectiveness in the cumulative scientific progress. In developing a platform for sharing research outputs, we present ModelHub.AI (www.modelhub.ai), a community-driven container-based software engine and platform for the structured dissemination of deep learning models. For contributors, the engine controls data flow throughout the inference cycle, while the contributor-facing standard template exposes model-specific functions including inference, as well as pre- and post-processing. Python and RESTful Application programming interfaces (APIs) enable users to interact with models hosted on ModelHub.AI and allows both researchers and developers to utilize models out-of-the-box. ModelHub.AI is domain-, data-, and framework-agnostic, catering to different workflows and contributors' preferences.
Repeatability of Multiparametric Prostate MRI Radiomics Features
Jul 16, 2018

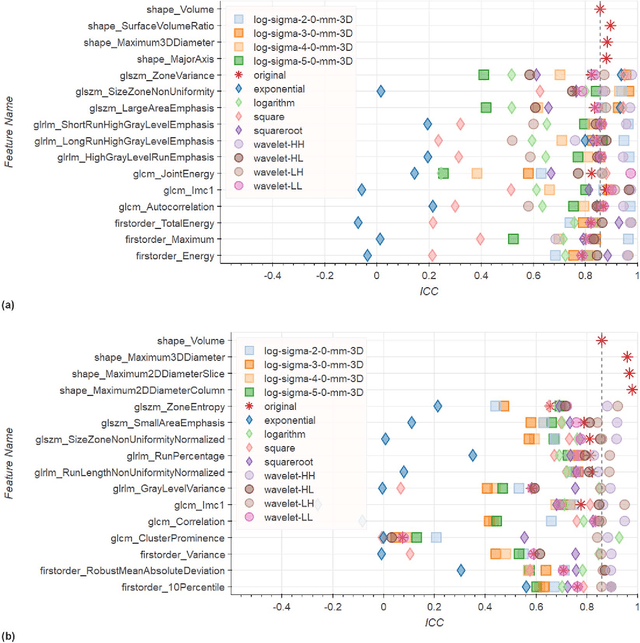
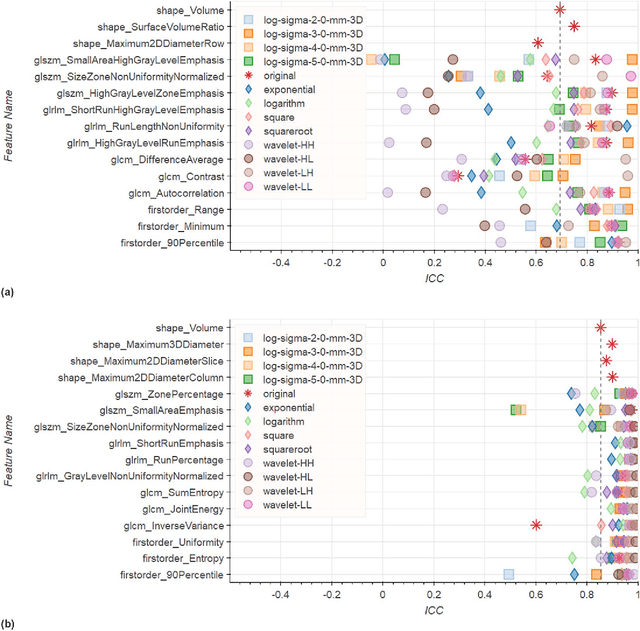
Abstract:In this study we assessed the repeatability of the values of radiomics features for small prostate tumors using test-retest? Multiparametric Magnetic Resonance Imaging (mpMRI) images. The premise of radiomics is that quantitative image features can serve as biomarkers characterizing disease. For such biomarkers to be useful, repeatability is a basic requirement, meaning its value must remain stable between two scans, if the conditions remain stable. We investigated repeatability of radiomics features under various preprocessing and extraction configurations including various image normalization schemes, different image pre-filtering, 2D vs 3D texture computation, and different bin widths for image discretization. Image registration as means to re-identify regions of interest across time points was evaluated against human-expert segmented regions in both time points. Even though we found many radiomics features and preprocessing combinations with a high repeatability (Intraclass Correlation Coefficient (ICC) > 0.85), our results indicate that overall the repeatability is highly sensitive to the processing parameters (under certain configurations, it can be below 0.0). Image normalization, using a variety of approaches considered, did not result in consistent improvements in repeatability. There was also no consistent improvement of repeatability through the use of pre-filtering options, or by using image registration between timepoints to improve consistency of the region of interest localization. Based on these results we urge caution when interpreting radiomics features and advise paying close attention to the processing configuration details of reported results. Furthermore, we advocate reporting all processing details in radiomics studies and strongly recommend making the implementation available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge