Sharon Peled
PSA-MIL: A Probabilistic Spatial Attention-Based Multiple Instance Learning for Whole Slide Image Classification
Mar 20, 2025Abstract:Whole Slide Images (WSIs) are high-resolution digital scans widely used in medical diagnostics. WSI classification is typically approached using Multiple Instance Learning (MIL), where the slide is partitioned into tiles treated as interconnected instances. While attention-based MIL methods aim to identify the most informative tiles, they often fail to fully exploit the spatial relationships among them, potentially overlooking intricate tissue structures crucial for accurate diagnosis. To address this limitation, we propose Probabilistic Spatial Attention MIL (PSA-MIL), a novel attention-based MIL framework that integrates spatial context into the attention mechanism through learnable distance-decayed priors, formulated within a probabilistic interpretation of self-attention as a posterior distribution. This formulation enables a dynamic inference of spatial relationships during training, eliminating the need for predefined assumptions often imposed by previous approaches. Additionally, we suggest a spatial pruning strategy for the posterior, effectively reducing self-attention's quadratic complexity. To further enhance spatial modeling, we introduce a diversity loss that encourages variation among attention heads, ensuring each captures distinct spatial representations. Together, PSA-MIL enables a more data-driven and adaptive integration of spatial context, moving beyond predefined constraints. We achieve state-of-the-art performance across both contextual and non-contextual baselines, while significantly reducing computational costs.
Multi-Cohort Framework with Cohort-Aware Attention and Adversarial Mutual-Information Minimization for Whole Slide Image Classification
Sep 17, 2024



Abstract:Whole Slide Images (WSIs) are critical for various clinical applications, including histopathological analysis. However, current deep learning approaches in this field predominantly focus on individual tumor types, limiting model generalization and scalability. This relatively narrow focus ultimately stems from the inherent heterogeneity in histopathology and the diverse morphological and molecular characteristics of different tumors. To this end, we propose a novel approach for multi-cohort WSI analysis, designed to leverage the diversity of different tumor types. We introduce a Cohort-Aware Attention module, enabling the capture of both shared and tumor-specific pathological patterns, enhancing cross-tumor generalization. Furthermore, we construct an adversarial cohort regularization mechanism to minimize cohort-specific biases through mutual information minimization. Additionally, we develop a hierarchical sample balancing strategy to mitigate cohort imbalances and promote unbiased learning. Together, these form a cohesive framework for unbiased multi-cohort WSI analysis. Extensive experiments on a uniquely constructed multi-cancer dataset demonstrate significant improvements in generalization, providing a scalable solution for WSI classification across diverse cancer types. Our code for the experiments is publicly available at <link>.
Repeatability of Multiparametric Prostate MRI Radiomics Features
Jul 16, 2018

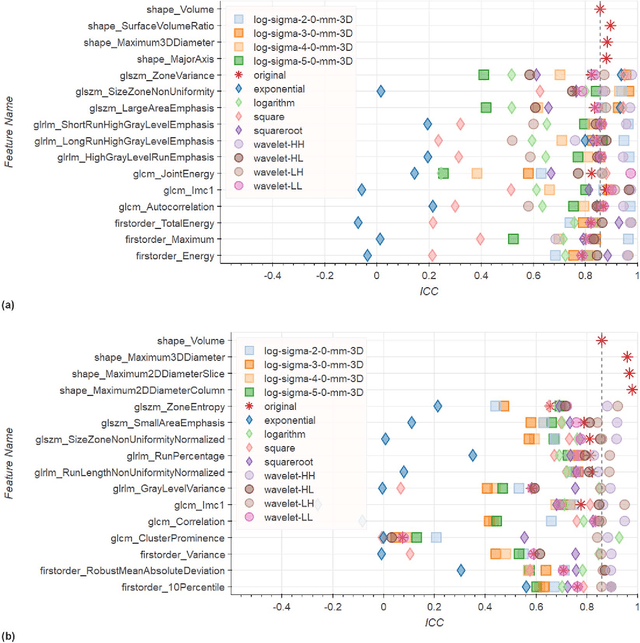
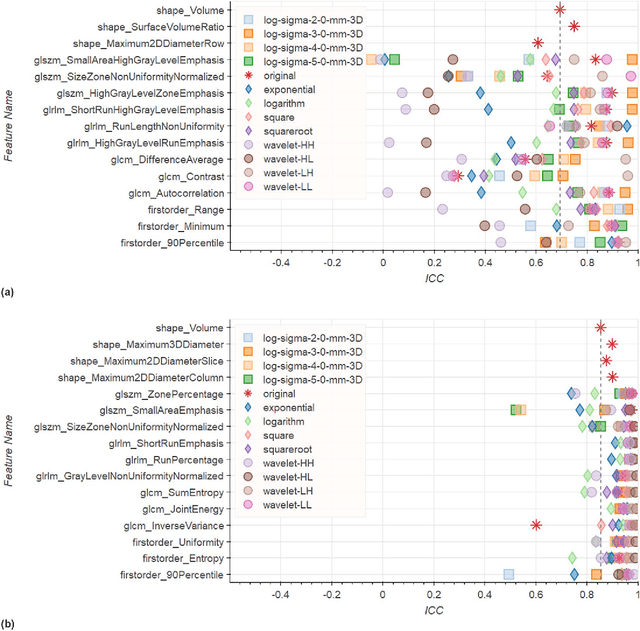
Abstract:In this study we assessed the repeatability of the values of radiomics features for small prostate tumors using test-retest? Multiparametric Magnetic Resonance Imaging (mpMRI) images. The premise of radiomics is that quantitative image features can serve as biomarkers characterizing disease. For such biomarkers to be useful, repeatability is a basic requirement, meaning its value must remain stable between two scans, if the conditions remain stable. We investigated repeatability of radiomics features under various preprocessing and extraction configurations including various image normalization schemes, different image pre-filtering, 2D vs 3D texture computation, and different bin widths for image discretization. Image registration as means to re-identify regions of interest across time points was evaluated against human-expert segmented regions in both time points. Even though we found many radiomics features and preprocessing combinations with a high repeatability (Intraclass Correlation Coefficient (ICC) > 0.85), our results indicate that overall the repeatability is highly sensitive to the processing parameters (under certain configurations, it can be below 0.0). Image normalization, using a variety of approaches considered, did not result in consistent improvements in repeatability. There was also no consistent improvement of repeatability through the use of pre-filtering options, or by using image registration between timepoints to improve consistency of the region of interest localization. Based on these results we urge caution when interpreting radiomics features and advise paying close attention to the processing configuration details of reported results. Furthermore, we advocate reporting all processing details in radiomics studies and strongly recommend making the implementation available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge