Moti Freiman
PSA-MIL: A Probabilistic Spatial Attention-Based Multiple Instance Learning for Whole Slide Image Classification
Mar 20, 2025Abstract:Whole Slide Images (WSIs) are high-resolution digital scans widely used in medical diagnostics. WSI classification is typically approached using Multiple Instance Learning (MIL), where the slide is partitioned into tiles treated as interconnected instances. While attention-based MIL methods aim to identify the most informative tiles, they often fail to fully exploit the spatial relationships among them, potentially overlooking intricate tissue structures crucial for accurate diagnosis. To address this limitation, we propose Probabilistic Spatial Attention MIL (PSA-MIL), a novel attention-based MIL framework that integrates spatial context into the attention mechanism through learnable distance-decayed priors, formulated within a probabilistic interpretation of self-attention as a posterior distribution. This formulation enables a dynamic inference of spatial relationships during training, eliminating the need for predefined assumptions often imposed by previous approaches. Additionally, we suggest a spatial pruning strategy for the posterior, effectively reducing self-attention's quadratic complexity. To further enhance spatial modeling, we introduce a diversity loss that encourages variation among attention heads, ensuring each captures distinct spatial representations. Together, PSA-MIL enables a more data-driven and adaptive integration of spatial context, moving beyond predefined constraints. We achieve state-of-the-art performance across both contextual and non-contextual baselines, while significantly reducing computational costs.
Enhancing Retinal Vessel Segmentation Generalization via Layout-Aware Generative Modelling
Mar 03, 2025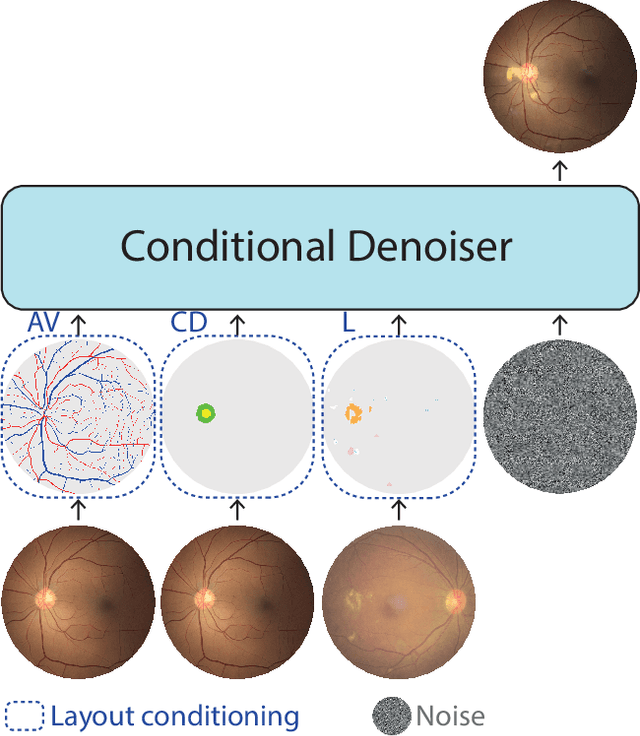
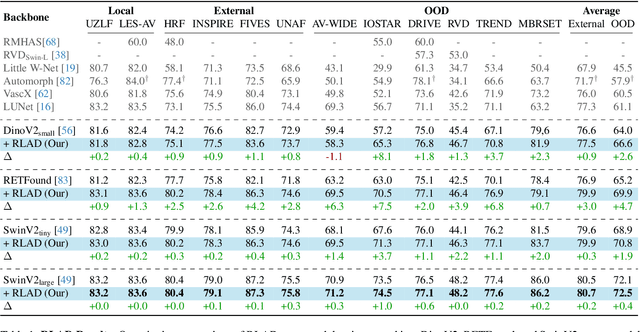
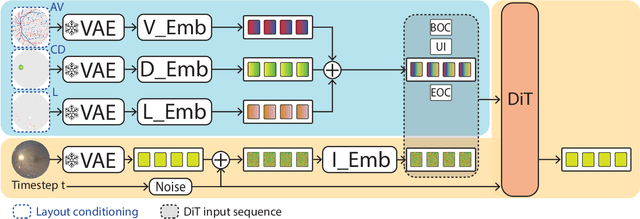
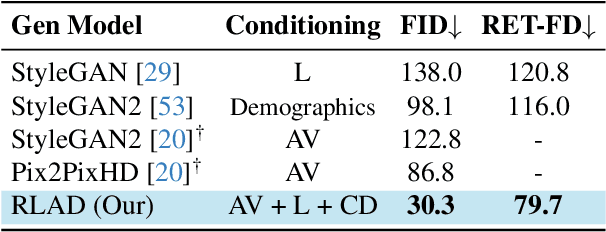
Abstract:Generalization in medical segmentation models is challenging due to limited annotated datasets and imaging variability. To address this, we propose Retinal Layout-Aware Diffusion (RLAD), a novel diffusion-based framework for generating controllable layout-aware images. RLAD conditions image generation on multiple key layout components extracted from real images, ensuring high structural fidelity while enabling diversity in other components. Applied to retinal fundus imaging, we augmented the training datasets by synthesizing paired retinal images and vessel segmentations conditioned on extracted blood vessels from real images, while varying other layout components such as lesions and the optic disc. Experiments demonstrated that RLAD-generated data improved generalization in retinal vessel segmentation by up to 8.1%. Furthermore, we present REYIA, a comprehensive dataset comprising 586 manually segmented retinal images. To foster reproducibility and drive innovation, both our code and dataset will be made publicly accessible.
T1-PILOT: Optimized Trajectories for T1 Mapping Acceleration
Feb 27, 2025Abstract:Cardiac T1 mapping provides critical quantitative insights into myocardial tissue composition, enabling the assessment of pathologies such as fibrosis, inflammation, and edema. However, the inherently dynamic nature of the heart imposes strict limits on acquisition times, making high-resolution T1 mapping a persistent challenge. Compressed sensing (CS) approaches have reduced scan durations by undersampling k-space and reconstructing images from partial data, and recent studies show that jointly optimizing the undersampling patterns with the reconstruction network can substantially improve performance. Still, most current T1 mapping pipelines rely on static, hand-crafted masks that do not exploit the full acceleration and accuracy potential. In this work, we introduce T1-PILOT: an end-to-end method that explicitly incorporates the T1 signal relaxation model into the sampling-reconstruction framework to guide the learning of non-Cartesian trajectories, crossframe alignment, and T1 decay estimation. Through extensive experiments on the CMRxRecon dataset, T1-PILOT significantly outperforms several baseline strategies (including learned single-mask and fixed radial or golden-angle sampling schemes), achieving higher T1 map fidelity at greater acceleration factors. In particular, we observe consistent gains in PSNR and VIF relative to existing methods, along with marked improvements in delineating finer myocardial structures. Our results highlight that optimizing sampling trajectories in tandem with the physical relaxation model leads to both enhanced quantitative accuracy and reduced acquisition times. Code for reproducing all results will be made publicly available upon publication.
Agent-Based Uncertainty Awareness Improves Automated Radiology Report Labeling with an Open-Source Large Language Model
Feb 02, 2025Abstract:Reliable extraction of structured data from radiology reports using Large Language Models (LLMs) remains challenging, especially for complex, non-English texts like Hebrew. This study introduces an agent-based uncertainty-aware approach to improve the trustworthiness of LLM predictions in medical applications. We analyzed 9,683 Hebrew radiology reports from Crohn's disease patients (from 2010 to 2023) across three medical centers. A subset of 512 reports was manually annotated for six gastrointestinal organs and 15 pathological findings, while the remaining reports were automatically annotated using HSMP-BERT. Structured data extraction was performed using Llama 3.1 (Llama 3-8b-instruct) with Bayesian Prompt Ensembles (BayesPE), which employed six semantically equivalent prompts to estimate uncertainty. An Agent-Based Decision Model integrated multiple prompt outputs into five confidence levels for calibrated uncertainty and was compared against three entropy-based models. Performance was evaluated using accuracy, F1 score, precision, recall, and Cohen's Kappa before and after filtering high-uncertainty cases. The agent-based model outperformed the baseline across all metrics, achieving an F1 score of 0.3967, recall of 0.6437, and Cohen's Kappa of 0.3006. After filtering high-uncertainty cases (greater than or equal to 0.5), the F1 score improved to 0.4787, and Kappa increased to 0.4258. Uncertainty histograms demonstrated clear separation between correct and incorrect predictions, with the agent-based model providing the most well-calibrated uncertainty estimates. By incorporating uncertainty-aware prompt ensembles and an agent-based decision model, this approach enhances the performance and reliability of LLMs in structured data extraction from radiology reports, offering a more interpretable and trustworthy solution for high-stakes medical applications.
Multi-Cohort Framework with Cohort-Aware Attention and Adversarial Mutual-Information Minimization for Whole Slide Image Classification
Sep 17, 2024



Abstract:Whole Slide Images (WSIs) are critical for various clinical applications, including histopathological analysis. However, current deep learning approaches in this field predominantly focus on individual tumor types, limiting model generalization and scalability. This relatively narrow focus ultimately stems from the inherent heterogeneity in histopathology and the diverse morphological and molecular characteristics of different tumors. To this end, we propose a novel approach for multi-cohort WSI analysis, designed to leverage the diversity of different tumor types. We introduce a Cohort-Aware Attention module, enabling the capture of both shared and tumor-specific pathological patterns, enhancing cross-tumor generalization. Furthermore, we construct an adversarial cohort regularization mechanism to minimize cohort-specific biases through mutual information minimization. Additionally, we develop a hierarchical sample balancing strategy to mitigate cohort imbalances and promote unbiased learning. Together, these form a cohesive framework for unbiased multi-cohort WSI analysis. Extensive experiments on a uniquely constructed multi-cancer dataset demonstrate significant improvements in generalization, providing a scalable solution for WSI classification across diverse cancer types. Our code for the experiments is publicly available at <link>.
SIMPLE: Simultaneous Multi-Plane Self-Supervised Learning for Isotropic MRI Restoration from Anisotropic Data
Aug 23, 2024Abstract:Magnetic resonance imaging (MRI) is crucial in diagnosing various abdominal conditions and anomalies. Traditional MRI scans often yield anisotropic data due to technical constraints, resulting in varying resolutions across spatial dimensions, which limits diagnostic accuracy and volumetric analysis. Super-resolution (SR) techniques aim to address these limitations by reconstructing isotropic high-resolution images from anisotropic data. However, current SR methods often rely on indirect mappings and limited training data, focusing mainly on two-dimensional improvements rather than achieving true three-dimensional isotropy. We introduce SIMPLE, a Simultaneous Multi-Plane Self-Supervised Learning approach for isotropic MRI restoration from anisotropic data. Our method leverages existing anisotropic clinical data acquired in different planes, bypassing the need for simulated downsampling processes. By considering the inherent three-dimensional nature of MRI data, SIMPLE ensures realistic isotropic data generation rather than solely improving through-plane slices. This approach flexibility allows it to be extended to multiple contrast types and acquisition methods commonly used in clinical settings. Our experiments show that SIMPLE outperforms state-of-the-art methods both quantitatively using the Kernel Inception Distance (KID) and semi-quantitatively through radiologist evaluations. The generated isotropic volume facilitates more accurate volumetric analysis and 3D reconstructions, promising significant improvements in clinical diagnostic capabilities.
MBSS-T1: Model-Based Self-Supervised Motion Correction for Robust Cardiac T1 Mapping
Aug 21, 2024Abstract:T1 mapping is a valuable quantitative MRI technique for diagnosing diffuse myocardial diseases. Traditional methods, relying on breath-hold sequences and echo triggering, face challenges with patient compliance and arrhythmias, limiting their effectiveness. Image registration can enable motion-robust T1 mapping, but inherent intensity differences between time points pose a challenge. We introduce MBSS-T1, a self-supervised model for motion correction in cardiac T1 mapping, constrained by physical and anatomical principles. The physical constraints ensure expected signal decay behavior, while the anatomical constraints maintain realistic deformations. The unique combination of these constraints ensures accurate T1 mapping along the longitudinal relaxation axis. MBSS-T1 outperformed baseline deep-learning-based image registration approaches in a 5-fold experiment on a public dataset of 210 patients (STONE sequence) and an internal dataset of 19 patients (MOLLI sequence). MBSS-T1 excelled in model fitting quality (R2: 0.974 vs. 0.941, 0.946), anatomical alignment (Dice score: 0.921 vs. 0.984, 0.988), and expert visual quality assessment for the presence of visible motion artifacts (4.33 vs. 3.34, 3.62). MBSS-T1 has the potential to enable motion-robust T1 mapping for a broader range of patients, overcoming challenges such as arrhythmias, and suboptimal compliance, and allowing for free-breathing T1 mapping without requiring large training datasets.
Leveraging Prompt-Learning for Structured Information Extraction from Crohn's Disease Radiology Reports in a Low-Resource Language
May 02, 2024Abstract:Automatic conversion of free-text radiology reports into structured data using Natural Language Processing (NLP) techniques is crucial for analyzing diseases on a large scale. While effective for tasks in widely spoken languages like English, generative large language models (LLMs) typically underperform with less common languages and can pose potential risks to patient privacy. Fine-tuning local NLP models is hindered by the skewed nature of real-world medical datasets, where rare findings represent a significant data imbalance. We introduce SMP-BERT, a novel prompt learning method that leverages the structured nature of reports to overcome these challenges. In our studies involving a substantial collection of Crohn's disease radiology reports in Hebrew (over 8,000 patients and 10,000 reports), SMP-BERT greatly surpassed traditional fine-tuning methods in performance, notably in detecting infrequent conditions (AUC: 0.99 vs 0.94, F1: 0.84 vs 0.34). SMP-BERT empowers more accurate AI diagnostics available for low-resource languages.
Automated Prediction of Breast Cancer Response to Neoadjuvant Chemotherapy from DWI Data
Apr 07, 2024Abstract:Effective surgical planning for breast cancer hinges on accurately predicting pathological complete response (pCR) to neoadjuvant chemotherapy (NAC). Diffusion-weighted MRI (DWI) and machine learning offer a non-invasive approach for early pCR assessment. However, most machine-learning models require manual tumor segmentation, a cumbersome and error-prone task. We propose a deep learning model employing "Size-Adaptive Lesion Weighting" for automatic DWI tumor segmentation to enhance pCR prediction accuracy. Despite histopathological changes during NAC complicating DWI image segmentation, our model demonstrates robust performance. Utilizing the BMMR2 challenge dataset, it matches human experts in pCR prediction pre-NAC with an area under the curve (AUC) of 0.76 vs. 0.796, and surpasses standard automated methods mid-NAC, with an AUC of 0.729 vs. 0.654 and 0.576. Our approach represents a significant advancement in automating breast cancer treatment planning, enabling more reliable pCR predictions without manual segmentation.
A self-attention model for robust rigid slice-to-volume registration of functional MRI
Apr 06, 2024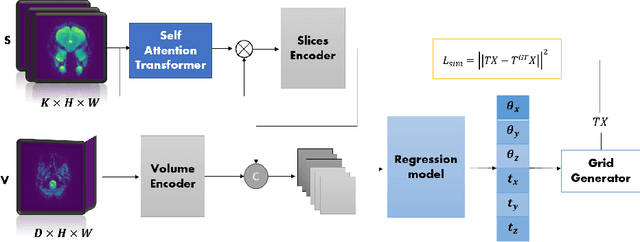
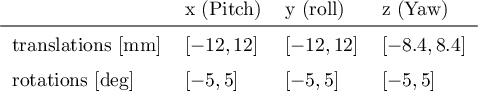
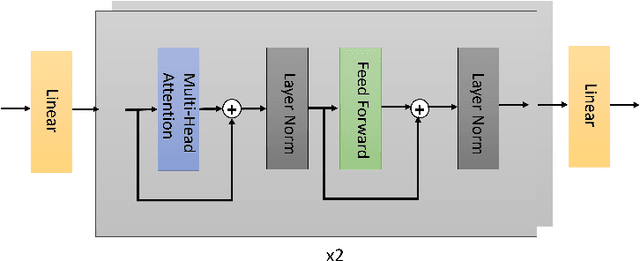
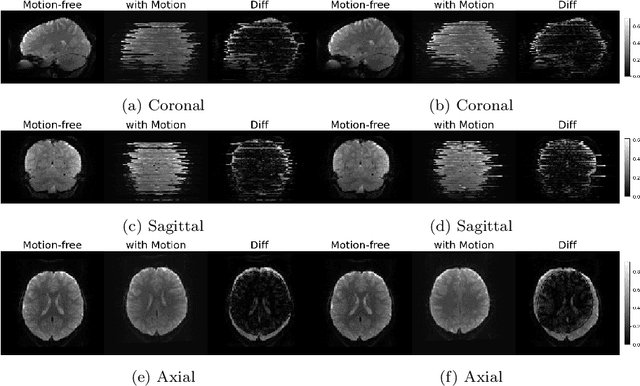
Abstract:Functional Magnetic Resonance Imaging (fMRI) is vital in neuroscience, enabling investigations into brain disorders, treatment monitoring, and brain function mapping. However, head motion during fMRI scans, occurring between shots of slice acquisition, can result in distortion, biased analyses, and increased costs due to the need for scan repetitions. Therefore, retrospective slice-level motion correction through slice-to-volume registration (SVR) is crucial. Previous studies have utilized deep learning (DL) based models to address the SVR task; however, they overlooked the uncertainty stemming from the input stack of slices and did not assign weighting or scoring to each slice. In this work, we introduce an end-to-end SVR model for aligning 2D fMRI slices with a 3D reference volume, incorporating a self-attention mechanism to enhance robustness against input data variations and uncertainties. It utilizes independent slice and volume encoders and a self-attention module to assign pixel-wise scores for each slice. We conducted evaluation experiments on 200 images involving synthetic rigid motion generated from 27 subjects belonging to the test set, from the publicly available Healthy Brain Network (HBN) dataset. Our experimental results demonstrate that our model achieves competitive performance in terms of alignment accuracy compared to state-of-the-art deep learning-based methods (Euclidean distance of $0.93$ [mm] vs. $1.86$ [mm]). Furthermore, our approach exhibits significantly faster registration speed compared to conventional iterative methods ($0.096$ sec. vs. $1.17$ sec.). Our end-to-end SVR model facilitates real-time head motion tracking during fMRI acquisition, ensuring reliability and robustness against uncertainties in inputs. source code, which includes the training and evaluations, will be available soon.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge