Hadas Ben-Atya
Agent-Based Uncertainty Awareness Improves Automated Radiology Report Labeling with an Open-Source Large Language Model
Feb 02, 2025Abstract:Reliable extraction of structured data from radiology reports using Large Language Models (LLMs) remains challenging, especially for complex, non-English texts like Hebrew. This study introduces an agent-based uncertainty-aware approach to improve the trustworthiness of LLM predictions in medical applications. We analyzed 9,683 Hebrew radiology reports from Crohn's disease patients (from 2010 to 2023) across three medical centers. A subset of 512 reports was manually annotated for six gastrointestinal organs and 15 pathological findings, while the remaining reports were automatically annotated using HSMP-BERT. Structured data extraction was performed using Llama 3.1 (Llama 3-8b-instruct) with Bayesian Prompt Ensembles (BayesPE), which employed six semantically equivalent prompts to estimate uncertainty. An Agent-Based Decision Model integrated multiple prompt outputs into five confidence levels for calibrated uncertainty and was compared against three entropy-based models. Performance was evaluated using accuracy, F1 score, precision, recall, and Cohen's Kappa before and after filtering high-uncertainty cases. The agent-based model outperformed the baseline across all metrics, achieving an F1 score of 0.3967, recall of 0.6437, and Cohen's Kappa of 0.3006. After filtering high-uncertainty cases (greater than or equal to 0.5), the F1 score improved to 0.4787, and Kappa increased to 0.4258. Uncertainty histograms demonstrated clear separation between correct and incorrect predictions, with the agent-based model providing the most well-calibrated uncertainty estimates. By incorporating uncertainty-aware prompt ensembles and an agent-based decision model, this approach enhances the performance and reliability of LLMs in structured data extraction from radiology reports, offering a more interpretable and trustworthy solution for high-stakes medical applications.
P2T2: a Physically-primed deep-neural-network approach for robust $T_{2}$ distribution estimation from quantitative $T_{2}$-weighted MRI
Dec 08, 2022Abstract:Estimation of the T2 distribution from multi-echo T2-Weighted MRI (T2W) data can provide insight into the microscopic content of tissue using macroscopic imaging. This information can be used as a biomarker for several pathologies, such as tumor characterization, osteoarthritis, and neurodegenerative diseases. Recently, deep neural network (DNN) based methods were proposed for T2 distribution estimation from MRI data. However, these methods are highly sensitive to distribution shifts such as variations in the echo-times (TE) used during acquisition. Therefore, DNN-based methods cannot be utilized in large-scale multi-institutional trials with heterogeneous acquisition protocols. We present P2T2, a new physically-primed DNN approach for T2 distribution estimation that is robust to different acquisition parameters while maintaining state-of-the-art estimation accuracy. Our P2T2 model encodes the forward model of the signal decay by taking as input the TE acquisition array, in addition to the MRI signal, and provides an estimate of the corresponding T2 distribution as its output. Our P2T2 model has improved the robustness against distribution shifts in the acquisition process by more than 50% compared to the previously proposed DNN model. When tested without any distribution shifts, our model achieved about the same accuracy. Finally, when applied to real human MRI data, our P2T2 model produced the most detailed Myelin-Water fraction maps compared to both the MIML model and classical approaches. Our proposed physically-primed approach improved the generalization capacity of DNN models for T2 distribution estimation and their robustness against distribution shifts compared to previous approaches without compromising the accuracy.
Non Parametric Data Augmentations Improve Deep-Learning based Brain Tumor Segmentation
Nov 25, 2021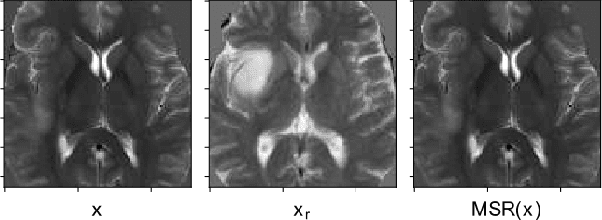
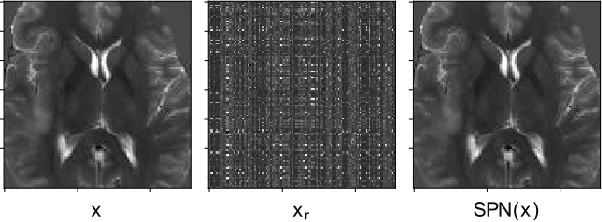
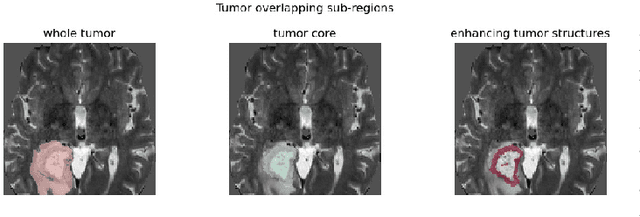
Abstract:Automatic brain tumor segmentation from Magnetic Resonance Imaging (MRI) data plays an important role in assessing tumor response to therapy and personalized treatment stratification.Manual segmentation is tedious and subjective.Deep-learning-based algorithms for brain tumor segmentation have the potential to provide objective and fast tumor segmentation.However, the training of such algorithms requires large datasets which are not always available. Data augmentation techniques may reduce the need for large datasets.However current approaches are mostly parametric and may result in suboptimal performance.We introduce two non-parametric methods of data augmentation for brain tumor segmentation: the mixed structure regularization (MSR) and shuffle pixels noise (SPN).We evaluated the added value of the MSR and SPN augmentation on the brain tumor segmentation (BraTS) 2018 challenge dataset with the encoder-decoder nnU-Net architecture as the segmentation algorithm.Both MSR and SPN improve the nnU-Net segmentation accuracy compared to parametric Gaussian noise augmentation.Mean dice score increased from 80% to 82% and p-values=0.0022, 0.0028 when comparing MSR to non-parametric augmentation for the tumor core and whole tumor experiments respectively.The proposed MSR and SPN augmentations have the potential to improve neural-networks performance in other tasks as well.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge