Jonathan Fhima
Benchmarking Ophthalmology Foundation Models for Clinically Significant Age Macular Degeneration Detection
May 08, 2025Abstract:Self-supervised learning (SSL) has enabled Vision Transformers (ViTs) to learn robust representations from large-scale natural image datasets, enhancing their generalization across domains. In retinal imaging, foundation models pretrained on either natural or ophthalmic data have shown promise, but the benefits of in-domain pretraining remain uncertain. To investigate this, we benchmark six SSL-pretrained ViTs on seven digital fundus image (DFI) datasets totaling 70,000 expert-annotated images for the task of moderate-to-late age-related macular degeneration (AMD) identification. Our results show that iBOT pretrained on natural images achieves the highest out-of-distribution generalization, with AUROCs of 0.80-0.97, outperforming domain-specific models, which achieved AUROCs of 0.78-0.96 and a baseline ViT-L with no pretraining, which achieved AUROCs of 0.68-0.91. These findings highlight the value of foundation models in improving AMD identification and challenge the assumption that in-domain pretraining is necessary. Furthermore, we release BRAMD, an open-access dataset (n=587) of DFIs with AMD labels from Brazil.
Enhancing Retinal Vessel Segmentation Generalization via Layout-Aware Generative Modelling
Mar 03, 2025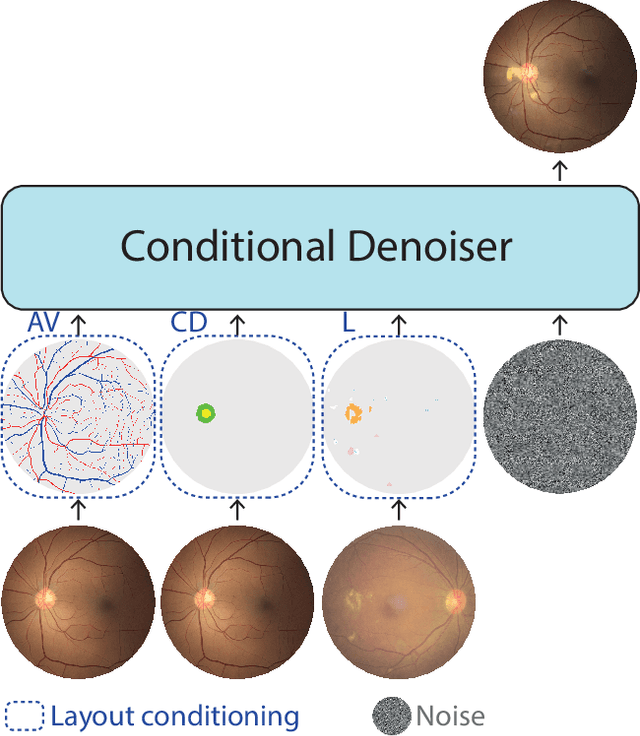
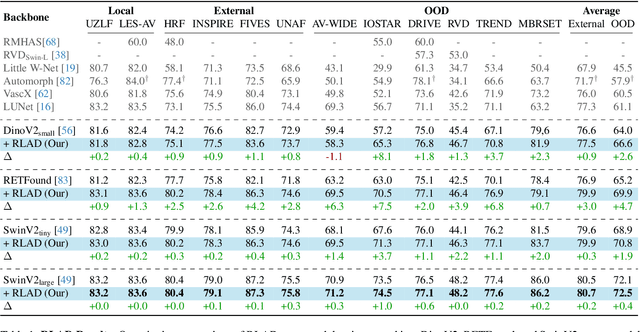
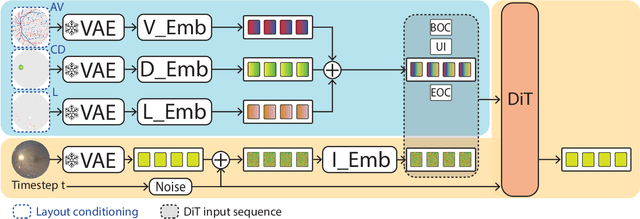
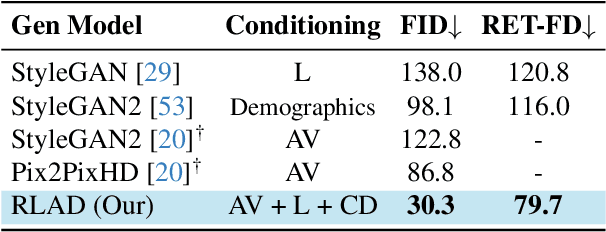
Abstract:Generalization in medical segmentation models is challenging due to limited annotated datasets and imaging variability. To address this, we propose Retinal Layout-Aware Diffusion (RLAD), a novel diffusion-based framework for generating controllable layout-aware images. RLAD conditions image generation on multiple key layout components extracted from real images, ensuring high structural fidelity while enabling diversity in other components. Applied to retinal fundus imaging, we augmented the training datasets by synthesizing paired retinal images and vessel segmentations conditioned on extracted blood vessels from real images, while varying other layout components such as lesions and the optic disc. Experiments demonstrated that RLAD-generated data improved generalization in retinal vessel segmentation by up to 8.1%. Furthermore, we present REYIA, a comprehensive dataset comprising 586 manually segmented retinal images. To foster reproducibility and drive innovation, both our code and dataset will be made publicly accessible.
GONet: A Generalizable Deep Learning Model for Glaucoma Detection
Feb 26, 2025



Abstract:Glaucomatous optic neuropathy (GON) is a prevalent ocular disease that can lead to irreversible vision loss if not detected early and treated. The traditional diagnostic approach for GON involves a set of ophthalmic examinations, which are time-consuming and require a visit to an ophthalmologist. Recent deep learning models for automating GON detection from digital fundus images (DFI) have shown promise but often suffer from limited generalizability across different ethnicities, disease groups and examination settings. To address these limitations, we introduce GONet, a robust deep learning model developed using seven independent datasets, including over 119,000 DFIs with gold-standard annotations and from patients of diverse geographic backgrounds. GONet consists of a DINOv2 pre-trained self-supervised vision transformers fine-tuned using a multisource domain strategy. GONet demonstrated high out-of-distribution generalizability, with an AUC of 0.85-0.99 in target domains. GONet performance was similar or superior to state-of-the-art works and was significantly superior to the cup-to-disc ratio, by up to 21.6%. GONet is available at [URL provided on publication]. We also contribute a new dataset consisting of 768 DFI with GON labels as open access.
TAP-VL: Text Layout-Aware Pre-training for Enriched Vision-Language Models
Nov 07, 2024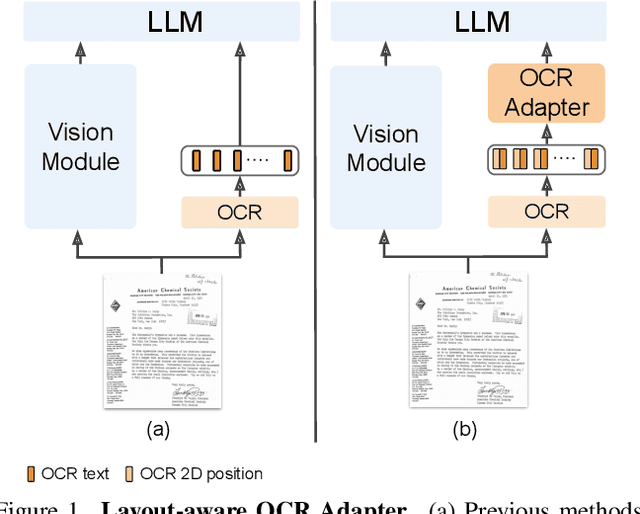
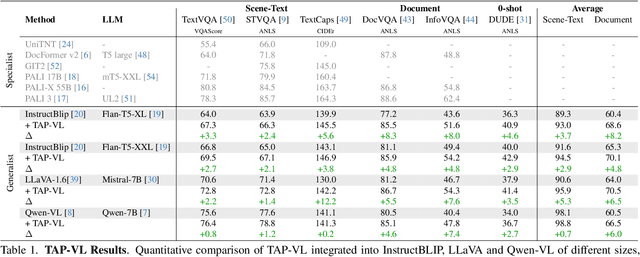
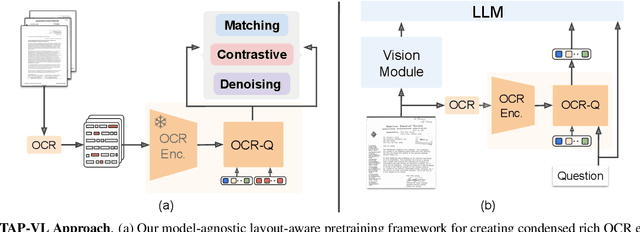
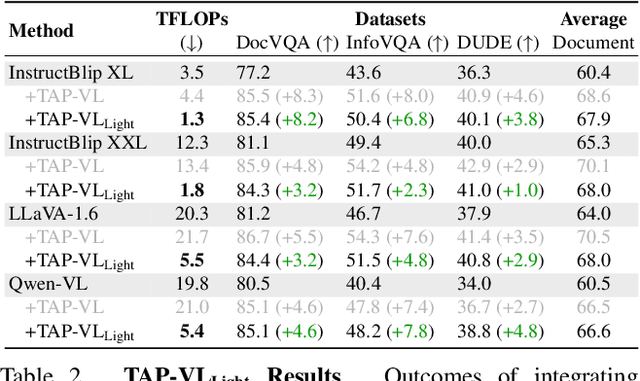
Abstract:Vision-Language (VL) models have garnered considerable research interest; however, they still face challenges in effectively handling text within images. To address this limitation, researchers have developed two approaches. The first method involves utilizing external Optical Character Recognition (OCR) tools to extract textual information from images, which is then prepended to other textual inputs. The second strategy focuses on employing extremely high-resolution images to improve text recognition capabilities. In this paper, we focus on enhancing the first strategy by introducing a novel method, named TAP-VL, which treats OCR information as a distinct modality and seamlessly integrates it into any VL model. TAP-VL employs a lightweight transformer-based OCR module to receive OCR with layout information, compressing it into a short fixed-length sequence for input into the LLM. Initially, we conduct model-agnostic pretraining of the OCR module on unlabeled documents, followed by its integration into any VL architecture through brief fine-tuning. Extensive experiments demonstrate consistent performance improvements when applying TAP-VL to top-performing VL models, across scene-text and document-based VL benchmarks.
DRStageNet: Deep Learning for Diabetic Retinopathy Staging from Fundus Images
Dec 22, 2023



Abstract:Diabetic retinopathy (DR) is a prevalent complication of diabetes associated with a significant risk of vision loss. Timely identification is critical to curb vision impairment. Algorithms for DR staging from digital fundus images (DFIs) have been recently proposed. However, models often fail to generalize due to distribution shifts between the source domain on which the model was trained and the target domain where it is deployed. A common and particularly challenging shift is often encountered when the source- and target-domain supports do not fully overlap. In this research, we introduce DRStageNet, a deep learning model designed to mitigate this challenge. We used seven publicly available datasets, comprising a total of 93,534 DFIs that cover a variety of patient demographics, ethnicities, geographic origins and comorbidities. We fine-tune DINOv2, a pretrained model of self-supervised vision transformer, and implement a multi-source domain fine-tuning strategy to enhance generalization performance. We benchmark and demonstrate the superiority of our method to two state-of-the-art benchmarks, including a recently published foundation model. We adapted the grad-rollout method to our regression task in order to provide high-resolution explainability heatmaps. The error analysis showed that 59\% of the main errors had incorrect reference labels. DRStageNet is accessible at URL [upon acceptance of the manuscript].
LUNet: Deep Learning for the Segmentation of Arterioles and Venules in High Resolution Fundus Images
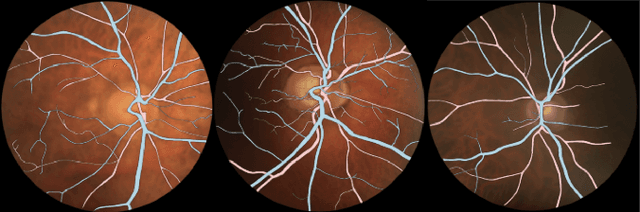
Sep 11, 2023Abstract:The retina is the only part of the human body in which blood vessels can be accessed non-invasively using imaging techniques such as digital fundus images (DFI). The spatial distribution of the retinal microvasculature may change with cardiovascular diseases and thus the eyes may be regarded as a window to our hearts. Computerized segmentation of the retinal arterioles and venules (A/V) is essential for automated microvasculature analysis. Using active learning, we created a new DFI dataset containing 240 crowd-sourced manual A/V segmentations performed by fifteen medical students and reviewed by an ophthalmologist, and developed LUNet, a novel deep learning architecture for high resolution A/V segmentation. LUNet architecture includes a double dilated convolutional block that aims to enhance the receptive field of the model and reduce its parameter count. Furthermore, LUNet has a long tail that operates at high resolution to refine the segmentation. The custom loss function emphasizes the continuity of the blood vessels. LUNet is shown to significantly outperform two state-of-the-art segmentation algorithms on the local test set as well as on four external test sets simulating distribution shifts across ethnicity, comorbidities, and annotators. We make the newly created dataset open access (upon publication).
Lirot.ai: A Novel Platform for Crowd-Sourcing Retinal Image Segmentations
Aug 22, 2022

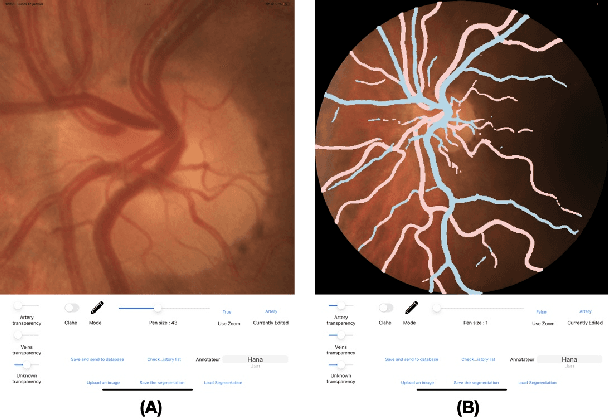
Abstract:Introduction: For supervised deep learning (DL) tasks, researchers need a large annotated dataset. In medical data science, one of the major limitations to develop DL models is the lack of annotated examples in large quantity. This is most often due to the time and expertise required to annotate. We introduce Lirot.ai, a novel platform for facilitating and crowd-sourcing image segmentations. Methods: Lirot.ai is composed of three components; an iPadOS client application named Lirot.ai-app, a backend server named Lirot.ai-server and a python API name Lirot.ai-API. Lirot.ai-app was developed in Swift 5.6 and Lirot.ai-server is a firebase backend. Lirot.ai-API allows the management of the database. Lirot.ai-app can be installed on as many iPadOS devices as needed so that annotators may be able to perform their segmentation simultaneously and remotely. We incorporate Apple Pencil compatibility, making the segmentation faster, more accurate, and more intuitive for the expert than any other computer-based alternative. Results: We demonstrate the usage of Lirot.ai for the creation of a retinal fundus dataset with reference vasculature segmentations. Discussion and future work: We will use active learning strategies to continue enlarging our retinal fundus dataset by including a more efficient process to select the images to be annotated and distribute them to annotators.
PVBM: A Python Vasculature Biomarker Toolbox Based On Retinal Blood Vessel Segmentation
Jul 31, 2022
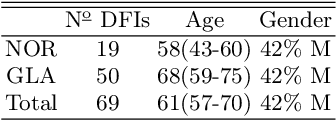
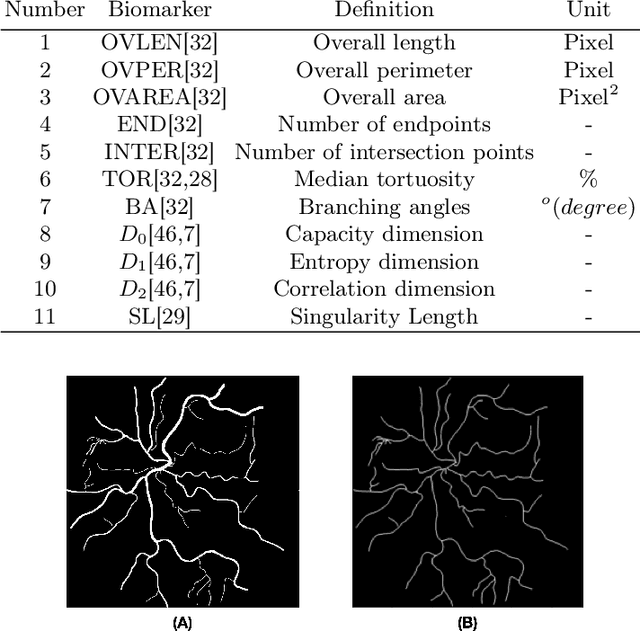
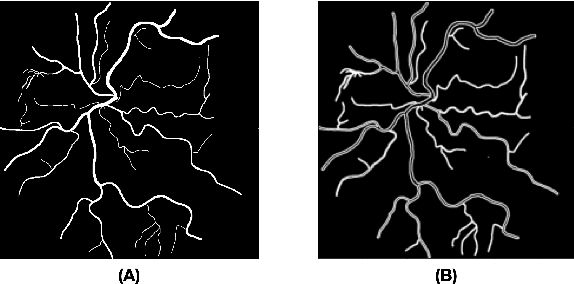
Abstract:Introduction: Blood vessels can be non-invasively visualized from a digital fundus image (DFI). Several studies have shown an association between cardiovascular risk and vascular features obtained from DFI. Recent advances in computer vision and image segmentation enable automatising DFI blood vessel segmentation. There is a need for a resource that can automatically compute digital vasculature biomarkers (VBM) from these segmented DFI. Methods: In this paper, we introduce a Python Vasculature BioMarker toolbox, denoted PVBM. A total of 11 VBMs were implemented. In particular, we introduce new algorithmic methods to estimate tortuosity and branching angles. Using PVBM, and as a proof of usability, we analyze geometric vascular differences between glaucomatous patients and healthy controls. Results: We built a fully automated vasculature biomarker toolbox based on DFI segmentations and provided a proof of usability to characterize the vascular changes in glaucoma. For arterioles and venules, all biomarkers were significant and lower in glaucoma patients compared to healthy controls except for tortuosity, venular singularity length and venular branching angles. Conclusion: We have automated the computation of 11 VBMs from retinal blood vessel segmentation. The PVBM toolbox is made open source under a GNU GPL 3 license and is available on physiozoo.com (following publication).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge