Joachim A. Behar
Modeling Day-Long ECG Signals to Predict Heart Failure Risk with Explainable AI
Dec 20, 2025Abstract:Heart failure (HF) affects 11.8% of adults aged 65 and older, reducing quality of life and longevity. Preventing HF can reduce morbidity and mortality. We hypothesized that artificial intelligence (AI) applied to 24-hour single-lead electrocardiogram (ECG) data could predict the risk of HF within five years. To research this, the Technion-Leumit Holter ECG (TLHE) dataset, including 69,663 recordings from 47,729 patients, collected over 20 years was used. Our deep learning model, DeepHHF, trained on 24-hour ECG recordings, achieved an area under the receiver operating characteristic curve of 0.80 that outperformed a model using 30-second segments and a clinical score. High-risk individuals identified by DeepHHF had a two-fold chance of hospitalization or death incidents. Explainability analysis showed DeepHHF focused on arrhythmias and heart abnormalities, with key attention between 8 AM and 3 PM. This study highlights the feasibility of deep learning to model 24-hour continuous ECG data, capturing paroxysmal events and circadian variations essential for reliable risk prediction. Artificial intelligence applied to single-lead Holter ECG is non-invasive, inexpensive, and widely accessible, making it a promising tool for HF risk prediction.
uPVC-Net: A Universal Premature Ventricular Contraction Detection Deep Learning Algorithm
Jun 12, 2025Abstract:Introduction: Premature Ventricular Contractions (PVCs) are common cardiac arrhythmias originating from the ventricles. Accurate detection remains challenging due to variability in electrocardiogram (ECG) waveforms caused by differences in lead placement, recording conditions, and population demographics. Methods: We developed uPVC-Net, a universal deep learning model to detect PVCs from any single-lead ECG recordings. The model is developed on four independent ECG datasets comprising a total of 8.3 million beats collected from Holter monitors and a modern wearable ECG patch. uPVC-Net employs a custom architecture and a multi-source, multi-lead training strategy. For each experiment, one dataset is held out to evaluate out-of-distribution (OOD) generalization. Results: uPVC-Net achieved an AUC between 97.8% and 99.1% on the held-out datasets. Notably, performance on wearable single-lead ECG data reached an AUC of 99.1%. Conclusion: uPVC-Net exhibits strong generalization across diverse lead configurations and populations, highlighting its potential for robust, real-world clinical deployment.
Benchmarking Ophthalmology Foundation Models for Clinically Significant Age Macular Degeneration Detection
May 08, 2025Abstract:Self-supervised learning (SSL) has enabled Vision Transformers (ViTs) to learn robust representations from large-scale natural image datasets, enhancing their generalization across domains. In retinal imaging, foundation models pretrained on either natural or ophthalmic data have shown promise, but the benefits of in-domain pretraining remain uncertain. To investigate this, we benchmark six SSL-pretrained ViTs on seven digital fundus image (DFI) datasets totaling 70,000 expert-annotated images for the task of moderate-to-late age-related macular degeneration (AMD) identification. Our results show that iBOT pretrained on natural images achieves the highest out-of-distribution generalization, with AUROCs of 0.80-0.97, outperforming domain-specific models, which achieved AUROCs of 0.78-0.96 and a baseline ViT-L with no pretraining, which achieved AUROCs of 0.68-0.91. These findings highlight the value of foundation models in improving AMD identification and challenge the assumption that in-domain pretraining is necessary. Furthermore, we release BRAMD, an open-access dataset (n=587) of DFIs with AMD labels from Brazil.
HYAMD High-Resolution Fundus Image Dataset for age related macular degeneration (AMD) Diagnosis
May 07, 2025

Abstract:The Hillel Yaffe Age Related Macular Degeneration (HYAMD) dataset is a longitudinal collection of 1,560 Digital Fundus Images (DFIs) from 325 patients examined at the Hillel Yaffe Medical Center (Hadera, Israel) between 2021 and 2024. The dataset includes an AMD cohort of 147 patients (aged 54-94) with varying stages of AMD and a control group of 190 diabetic retinopathy (DR) patients (aged 24-92). AMD diagnoses were based on comprehensive clinical ophthalmic evaluations, supported by Optical Coherence Tomography (OCT) and OCT angiography. Non-AMD DFIs were sourced from DR patients without concurrent AMD, diagnosed using macular OCT, fluorescein angiography, and widefield imaging. HYAMD provides gold-standard annotations, ensuring AMD labels were assigned following a full clinical assessment. Images were captured with a DRI OCT Triton (Topcon) camera, offering a 45 deg field of view and 1960 x 1934 pixel resolution. To the best of our knowledge, HYAMD is the first open-access retinal dataset from an Israeli sample, designed to support AMD identification using machine learning models.
Enhancing Retinal Vessel Segmentation Generalization via Layout-Aware Generative Modelling
Mar 03, 2025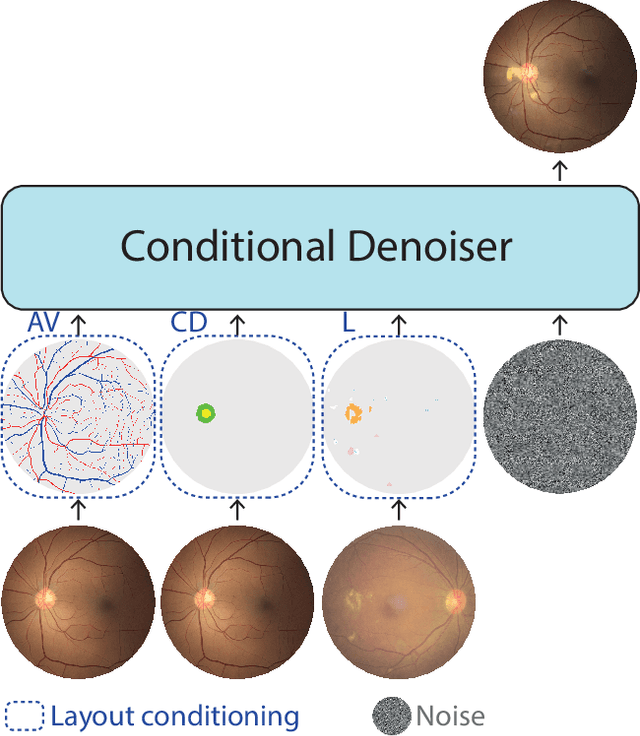
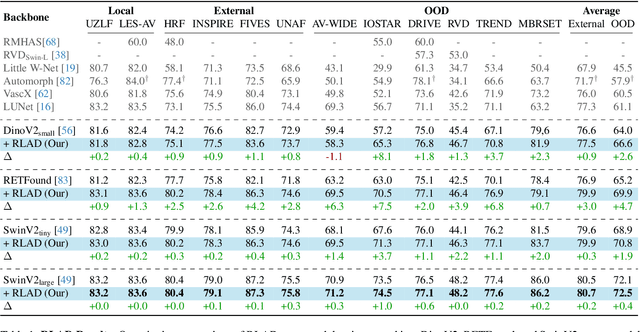
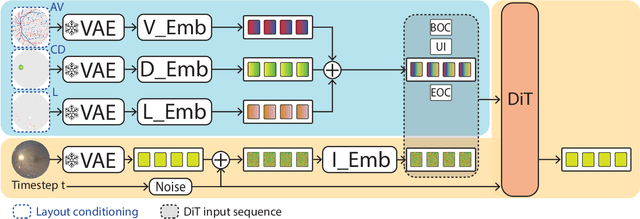
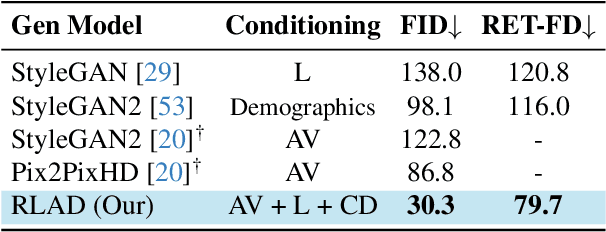
Abstract:Generalization in medical segmentation models is challenging due to limited annotated datasets and imaging variability. To address this, we propose Retinal Layout-Aware Diffusion (RLAD), a novel diffusion-based framework for generating controllable layout-aware images. RLAD conditions image generation on multiple key layout components extracted from real images, ensuring high structural fidelity while enabling diversity in other components. Applied to retinal fundus imaging, we augmented the training datasets by synthesizing paired retinal images and vessel segmentations conditioned on extracted blood vessels from real images, while varying other layout components such as lesions and the optic disc. Experiments demonstrated that RLAD-generated data improved generalization in retinal vessel segmentation by up to 8.1%. Furthermore, we present REYIA, a comprehensive dataset comprising 586 manually segmented retinal images. To foster reproducibility and drive innovation, both our code and dataset will be made publicly accessible.
GONet: A Generalizable Deep Learning Model for Glaucoma Detection
Feb 26, 2025



Abstract:Glaucomatous optic neuropathy (GON) is a prevalent ocular disease that can lead to irreversible vision loss if not detected early and treated. The traditional diagnostic approach for GON involves a set of ophthalmic examinations, which are time-consuming and require a visit to an ophthalmologist. Recent deep learning models for automating GON detection from digital fundus images (DFI) have shown promise but often suffer from limited generalizability across different ethnicities, disease groups and examination settings. To address these limitations, we introduce GONet, a robust deep learning model developed using seven independent datasets, including over 119,000 DFIs with gold-standard annotations and from patients of diverse geographic backgrounds. GONet consists of a DINOv2 pre-trained self-supervised vision transformers fine-tuned using a multisource domain strategy. GONet demonstrated high out-of-distribution generalizability, with an AUC of 0.85-0.99 in target domains. GONet performance was similar or superior to state-of-the-art works and was significantly superior to the cup-to-disc ratio, by up to 21.6%. GONet is available at [URL provided on publication]. We also contribute a new dataset consisting of 768 DFI with GON labels as open access.
SHDB-AF: a Japanese Holter ECG database of atrial fibrillation
Jun 22, 2024Abstract:Atrial fibrillation (AF) is a common atrial arrhythmia that impairs quality of life and causes embolic stroke, heart failure and other complications. Recent advancements in machine learning (ML) and deep learning (DL) have shown potential for enhancing diagnostic accuracy. It is essential for DL models to be robust and generalizable across variations in ethnicity, age, sex, and other factors. Although a number of ECG database have been made available to the research community, none includes a Japanese population sample. Saitama Heart Database Atrial Fibrillation (SHDB-AF) is a novel open-sourced Holter ECG database from Japan, containing data from 100 unique patients with paroxysmal AF. Each record in SHDB-AF is 24 hours long and sampled at 200 Hz, totaling 24 million seconds of ECG data.
SleepPPG-Net2: Deep learning generalization for sleep staging from photoplethysmography
Apr 10, 2024



Abstract:Background: Sleep staging is a fundamental component in the diagnosis of sleep disorders and the management of sleep health. Traditionally, this analysis is conducted in clinical settings and involves a time-consuming scoring procedure. Recent data-driven algorithms for sleep staging, using the photoplethysmogram (PPG) time series, have shown high performance on local test sets but lower performance on external datasets due to data drift. Methods: This study aimed to develop a generalizable deep learning model for the task of four class (wake, light, deep, and rapid eye movement (REM)) sleep staging from raw PPG physiological time-series. Six sleep datasets, totaling 2,574 patients recordings, were used. In order to create a more generalizable representation, we developed and evaluated a deep learning model called SleepPPG-Net2, which employs a multi-source domain training approach.SleepPPG-Net2 was benchmarked against two state-of-the-art models. Results: SleepPPG-Net2 showed consistently higher performance over benchmark approaches, with generalization performance (Cohen's kappa) improving by up to 19%. Performance disparities were observed in relation to age, sex, and sleep apnea severity. Conclusion: SleepPPG-Net2 sets a new standard for staging sleep from raw PPG time-series.
RawECGNet: Deep Learning Generalization for Atrial Fibrillation Detection from the Raw ECG
Dec 26, 2023



Abstract:Introduction: Deep learning models for detecting episodes of atrial fibrillation (AF) using rhythm information in long-term, ambulatory ECG recordings have shown high performance. However, the rhythm-based approach does not take advantage of the morphological information conveyed by the different ECG waveforms, particularly the f-waves. As a result, the performance of such models may be inherently limited. Methods: To address this limitation, we have developed a deep learning model, named RawECGNet, to detect episodes of AF and atrial flutter (AFl) using the raw, single-lead ECG. We compare the generalization performance of RawECGNet on two external data sets that account for distribution shifts in geography, ethnicity, and lead position. RawECGNet is further benchmarked against a state-of-the-art deep learning model, named ArNet2, which utilizes rhythm information as input. Results: Using RawECGNet, the results for the different leads in the external test sets in terms of the F1 score were 0.91--0.94 in RBDB and 0.93 in SHDB, compared to 0.89--0.91 in RBDB and 0.91 in SHDB for ArNet2. The results highlight RawECGNet as a high-performance, generalizable algorithm for detection of AF and AFl episodes, exploiting information on both rhythm and morphology.
DRStageNet: Deep Learning for Diabetic Retinopathy Staging from Fundus Images
Dec 22, 2023



Abstract:Diabetic retinopathy (DR) is a prevalent complication of diabetes associated with a significant risk of vision loss. Timely identification is critical to curb vision impairment. Algorithms for DR staging from digital fundus images (DFIs) have been recently proposed. However, models often fail to generalize due to distribution shifts between the source domain on which the model was trained and the target domain where it is deployed. A common and particularly challenging shift is often encountered when the source- and target-domain supports do not fully overlap. In this research, we introduce DRStageNet, a deep learning model designed to mitigate this challenge. We used seven publicly available datasets, comprising a total of 93,534 DFIs that cover a variety of patient demographics, ethnicities, geographic origins and comorbidities. We fine-tune DINOv2, a pretrained model of self-supervised vision transformer, and implement a multi-source domain fine-tuning strategy to enhance generalization performance. We benchmark and demonstrate the superiority of our method to two state-of-the-art benchmarks, including a recently published foundation model. We adapted the grad-rollout method to our regression task in order to provide high-resolution explainability heatmaps. The error analysis showed that 59\% of the main errors had incorrect reference labels. DRStageNet is accessible at URL [upon acceptance of the manuscript].
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge