Yosef E. Maruvka
PSA-MIL: A Probabilistic Spatial Attention-Based Multiple Instance Learning for Whole Slide Image Classification
Mar 20, 2025Abstract:Whole Slide Images (WSIs) are high-resolution digital scans widely used in medical diagnostics. WSI classification is typically approached using Multiple Instance Learning (MIL), where the slide is partitioned into tiles treated as interconnected instances. While attention-based MIL methods aim to identify the most informative tiles, they often fail to fully exploit the spatial relationships among them, potentially overlooking intricate tissue structures crucial for accurate diagnosis. To address this limitation, we propose Probabilistic Spatial Attention MIL (PSA-MIL), a novel attention-based MIL framework that integrates spatial context into the attention mechanism through learnable distance-decayed priors, formulated within a probabilistic interpretation of self-attention as a posterior distribution. This formulation enables a dynamic inference of spatial relationships during training, eliminating the need for predefined assumptions often imposed by previous approaches. Additionally, we suggest a spatial pruning strategy for the posterior, effectively reducing self-attention's quadratic complexity. To further enhance spatial modeling, we introduce a diversity loss that encourages variation among attention heads, ensuring each captures distinct spatial representations. Together, PSA-MIL enables a more data-driven and adaptive integration of spatial context, moving beyond predefined constraints. We achieve state-of-the-art performance across both contextual and non-contextual baselines, while significantly reducing computational costs.
Multi-Cohort Framework with Cohort-Aware Attention and Adversarial Mutual-Information Minimization for Whole Slide Image Classification
Sep 17, 2024



Abstract:Whole Slide Images (WSIs) are critical for various clinical applications, including histopathological analysis. However, current deep learning approaches in this field predominantly focus on individual tumor types, limiting model generalization and scalability. This relatively narrow focus ultimately stems from the inherent heterogeneity in histopathology and the diverse morphological and molecular characteristics of different tumors. To this end, we propose a novel approach for multi-cohort WSI analysis, designed to leverage the diversity of different tumor types. We introduce a Cohort-Aware Attention module, enabling the capture of both shared and tumor-specific pathological patterns, enhancing cross-tumor generalization. Furthermore, we construct an adversarial cohort regularization mechanism to minimize cohort-specific biases through mutual information minimization. Additionally, we develop a hierarchical sample balancing strategy to mitigate cohort imbalances and promote unbiased learning. Together, these form a cohesive framework for unbiased multi-cohort WSI analysis. Extensive experiments on a uniquely constructed multi-cancer dataset demonstrate significant improvements in generalization, providing a scalable solution for WSI classification across diverse cancer types. Our code for the experiments is publicly available at <link>.
CIMIL-CRC: a clinically-informed multiple instance learning framework for patient-level colorectal cancer molecular subtypes classification from H\&E stained images
Jan 29, 2024Abstract:Treatment approaches for colorectal cancer (CRC) are highly dependent on the molecular subtype, as immunotherapy has shown efficacy in cases with microsatellite instability (MSI) but is ineffective for the microsatellite stable (MSS) subtype. There is promising potential in utilizing deep neural networks (DNNs) to automate the differentiation of CRC subtypes by analyzing Hematoxylin and Eosin (H\&E) stained whole-slide images (WSIs). Due to the extensive size of WSIs, Multiple Instance Learning (MIL) techniques are typically explored. However, existing MIL methods focus on identifying the most representative image patches for classification, which may result in the loss of critical information. Additionally, these methods often overlook clinically relevant information, like the tendency for MSI class tumors to predominantly occur on the proximal (right side) colon. We introduce `CIMIL-CRC', a DNN framework that: 1) solves the MSI/MSS MIL problem by efficiently combining a pre-trained feature extraction model with principal component analysis (PCA) to aggregate information from all patches, and 2) integrates clinical priors, particularly the tumor location within the colon, into the model to enhance patient-level classification accuracy. We assessed our CIMIL-CRC method using the average area under the curve (AUC) from a 5-fold cross-validation experimental setup for model development on the TCGA-CRC-DX cohort, contrasting it with a baseline patch-level classification, MIL-only approach, and Clinically-informed patch-level classification approach. Our CIMIL-CRC outperformed all methods (AUROC: $0.92\pm0.002$ (95\% CI 0.91-0.92), vs. $0.79\pm0.02$ (95\% CI 0.76-0.82), $0.86\pm0.01$ (95\% CI 0.85-0.88), and $0.87\pm0.01$ (95\% CI 0.86-0.88), respectively). The improvement was statistically significant.
Biologically-primed deep neural network improves colorectal Cancer Molecular subtypes prediction from H&E stained images
Mar 26, 2023Abstract:Colorectal cancer (CRC) molecular subtypes play a crucial role in determining treatment options. Immunotherapy is effective for the microsatellite instability (MSI) subtype of CRC, but not for the microsatellite stability (MSS) subtype. Recently, convolutional neural networks (CNNs) have been proposed for automated determination of CRC subtypes from H\&E stained histopathological images. However, previous CNN architectures only consider binary outcomes of MSI or MSS, and do not account for additional biological cues that may affect the histopathological imaging phenotype. In this study, we propose a biologically-primed CNN (BP-CNN) architecture for CRC subtype classification from H\&E stained images. Our BP-CNN accounts for additional biological cues by casting the binary classification outcome into a biologically-informed multi-class outcome. We evaluated the BP-CNN approach using a 5-fold cross-validation experimental setup for model development on the TCGA-CRC-DX cohort, comparing it to a baseline binary classification CNN. Our BP-CNN achieved superior performance when using either single-nucleotide-polymorphism (SNP) molecular features (AUC: 0.824$\pm$0.02 vs. 0.761$\pm$0.04, paired t-test, p$<$0.05) or CpG-Island methylation phenotype (CIMP) molecular features (AUC: 0.834$\pm$0.01 vs. 0.787$\pm$0.03, paired t-test, p$<$0.05). A combination of CIMP and SNP models further improved classification accuracy (AUC: 0.847$\pm$0.01 vs. 0.787$\pm$0.03, paired t-test, p$=$0.01). Our BP-CNN approach has the potential to provide insight into the biological cues that influence cancer histopathological imaging phenotypes and to improve the accuracy of deep-learning-based methods for determining cancer subtypes from histopathological imaging data.
Patient-level Microsatellite Stability Assessment from Whole Slide Images By Combining Momentum Contrast Learning and Group Patch Embeddings
Aug 22, 2022
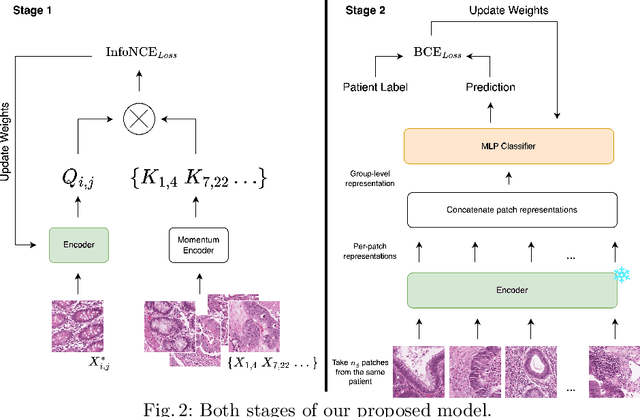
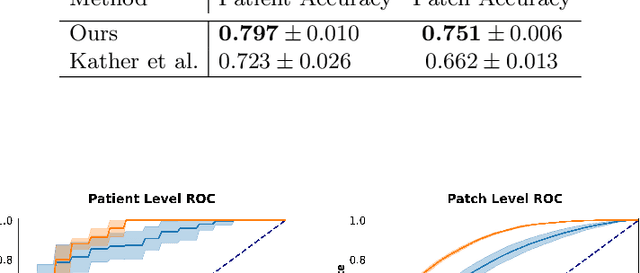
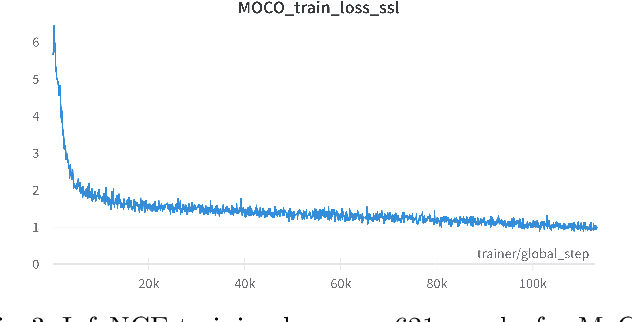
Abstract:Assessing microsatellite stability status of a patient's colorectal cancer is crucial in personalizing treatment regime. Recently, convolutional-neural-networks (CNN) combined with transfer-learning approaches were proposed to circumvent traditional laboratory testing for determining microsatellite status from hematoxylin and eosin stained biopsy whole slide images (WSI). However, the high resolution of WSI practically prevent direct classification of the entire WSI. Current approaches bypass the WSI high resolution by first classifying small patches extracted from the WSI, and then aggregating patch-level classification logits to deduce the patient-level status. Such approaches limit the capacity to capture important information which resides at the high resolution WSI data. We introduce an effective approach to leverage WSI high resolution information by momentum contrastive learning of patch embeddings along with training a patient-level classifier on groups of those embeddings. Our approach achieves up to 7.4\% better accuracy compared to the straightforward patch-level classification and patient level aggregation approach with a higher stability (AUC, $0.91 \pm 0.01$ vs. $0.85 \pm 0.04$, p-value$<0.01$). Our code can be found at https://github.com/TechnionComputationalMRILab/colorectal_cancer_ai.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge