Patient-level Microsatellite Stability Assessment from Whole Slide Images By Combining Momentum Contrast Learning and Group Patch Embeddings
Paper and Code
Aug 22, 2022
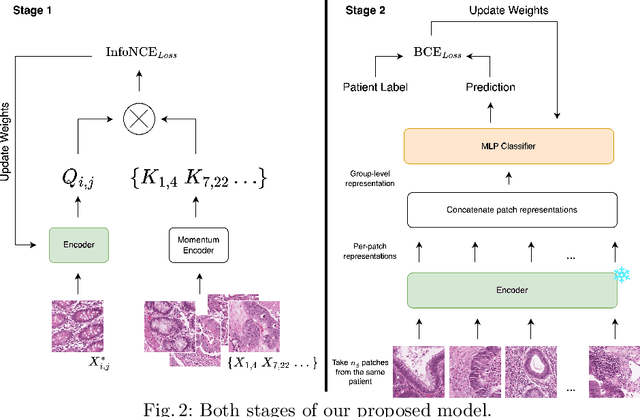
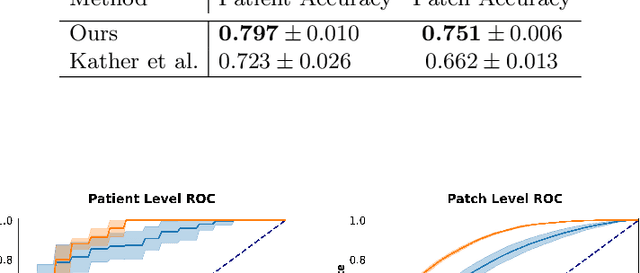
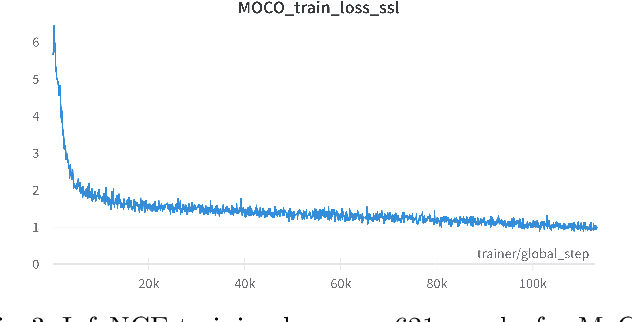
Assessing microsatellite stability status of a patient's colorectal cancer is crucial in personalizing treatment regime. Recently, convolutional-neural-networks (CNN) combined with transfer-learning approaches were proposed to circumvent traditional laboratory testing for determining microsatellite status from hematoxylin and eosin stained biopsy whole slide images (WSI). However, the high resolution of WSI practically prevent direct classification of the entire WSI. Current approaches bypass the WSI high resolution by first classifying small patches extracted from the WSI, and then aggregating patch-level classification logits to deduce the patient-level status. Such approaches limit the capacity to capture important information which resides at the high resolution WSI data. We introduce an effective approach to leverage WSI high resolution information by momentum contrastive learning of patch embeddings along with training a patient-level classifier on groups of those embeddings. Our approach achieves up to 7.4\% better accuracy compared to the straightforward patch-level classification and patient level aggregation approach with a higher stability (AUC, $0.91 \pm 0.01$ vs. $0.85 \pm 0.04$, p-value$<0.01$). Our code can be found at https://github.com/TechnionComputationalMRILab/colorectal_cancer_ai.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge