Daniel Shats
A large dataset curation and benchmark for drug target interaction
Jan 30, 2024Abstract:Bioactivity data plays a key role in drug discovery and repurposing. The resource-demanding nature of \textit{in vitro} and \textit{in vivo} experiments, as well as the recent advances in data-driven computational biochemistry research, highlight the importance of \textit{in silico} drug target interaction (DTI) prediction approaches. While numerous large public bioactivity data sources exist, research in the field could benefit from better standardization of existing data resources. At present, different research works that share similar goals are often difficult to compare properly because of different choices of data sources and train/validation/test split strategies. Additionally, many works are based on small data subsets, leading to results and insights of possible limited validity. In this paper we propose a way to standardize and represent efficiently a very large dataset curated from multiple public sources, split the data into train, validation and test sets based on different meaningful strategies, and provide a concrete evaluation protocol to accomplish a benchmark. We analyze the proposed data curation, prove its usefulness and validate the proposed benchmark through experimental studies based on an existing neural network model.
The KiTS21 Challenge: Automatic segmentation of kidneys, renal tumors, and renal cysts in corticomedullary-phase CT
Jul 05, 2023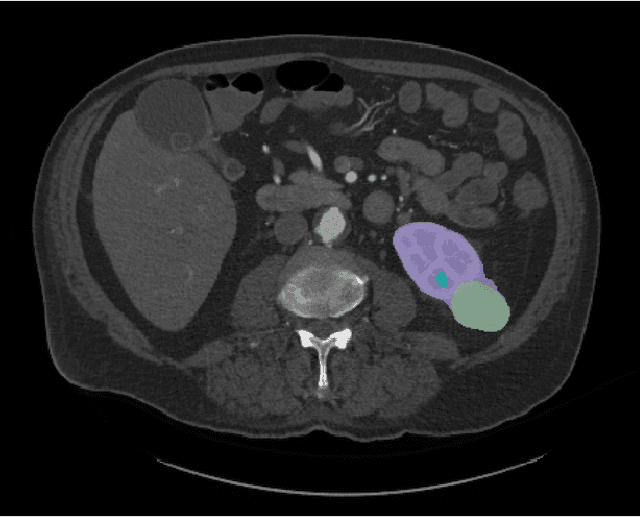
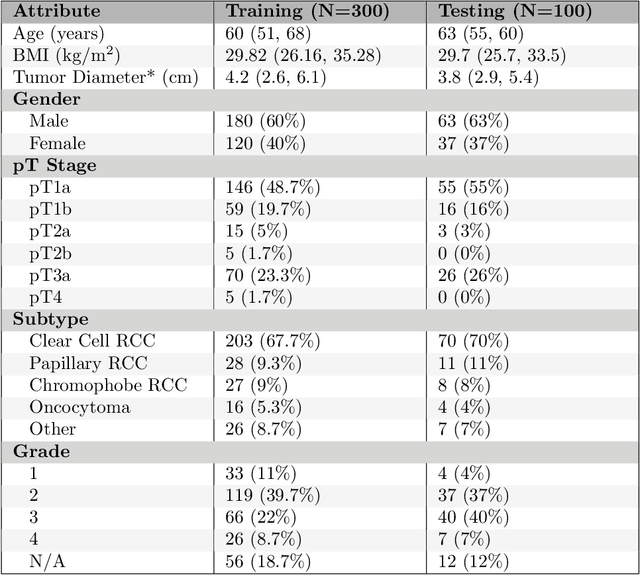
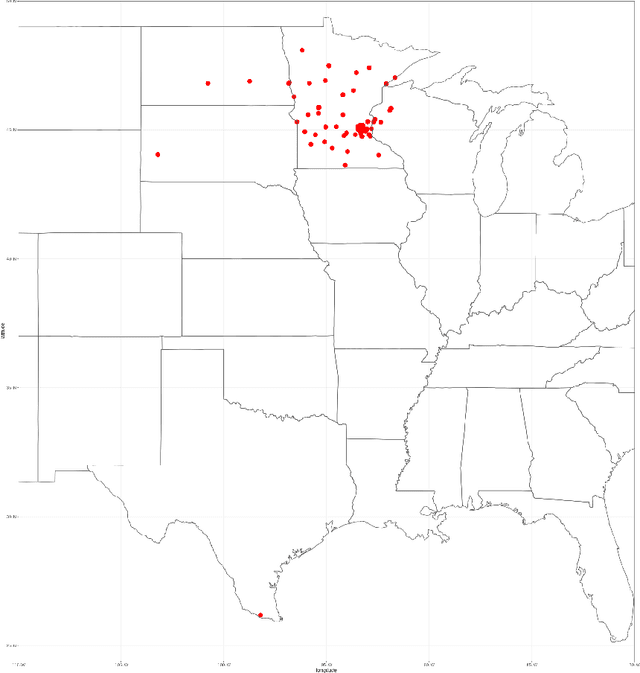
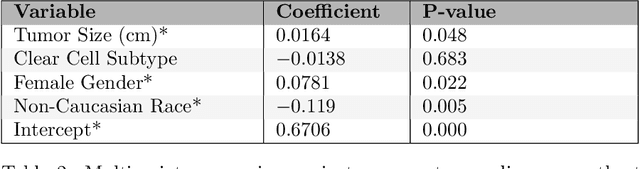
Abstract:This paper presents the challenge report for the 2021 Kidney and Kidney Tumor Segmentation Challenge (KiTS21) held in conjunction with the 2021 international conference on Medical Image Computing and Computer Assisted Interventions (MICCAI). KiTS21 is a sequel to its first edition in 2019, and it features a variety of innovations in how the challenge was designed, in addition to a larger dataset. A novel annotation method was used to collect three separate annotations for each region of interest, and these annotations were performed in a fully transparent setting using a web-based annotation tool. Further, the KiTS21 test set was collected from an outside institution, challenging participants to develop methods that generalize well to new populations. Nonetheless, the top-performing teams achieved a significant improvement over the state of the art set in 2019, and this performance is shown to inch ever closer to human-level performance. An in-depth meta-analysis is presented describing which methods were used and how they faired on the leaderboard, as well as the characteristics of which cases generally saw good performance, and which did not. Overall KiTS21 facilitated a significant advancement in the state of the art in kidney tumor segmentation, and provides useful insights that are applicable to the field of semantic segmentation as a whole.
Biologically-primed deep neural network improves colorectal Cancer Molecular subtypes prediction from H&E stained images
Mar 26, 2023Abstract:Colorectal cancer (CRC) molecular subtypes play a crucial role in determining treatment options. Immunotherapy is effective for the microsatellite instability (MSI) subtype of CRC, but not for the microsatellite stability (MSS) subtype. Recently, convolutional neural networks (CNNs) have been proposed for automated determination of CRC subtypes from H\&E stained histopathological images. However, previous CNN architectures only consider binary outcomes of MSI or MSS, and do not account for additional biological cues that may affect the histopathological imaging phenotype. In this study, we propose a biologically-primed CNN (BP-CNN) architecture for CRC subtype classification from H\&E stained images. Our BP-CNN accounts for additional biological cues by casting the binary classification outcome into a biologically-informed multi-class outcome. We evaluated the BP-CNN approach using a 5-fold cross-validation experimental setup for model development on the TCGA-CRC-DX cohort, comparing it to a baseline binary classification CNN. Our BP-CNN achieved superior performance when using either single-nucleotide-polymorphism (SNP) molecular features (AUC: 0.824$\pm$0.02 vs. 0.761$\pm$0.04, paired t-test, p$<$0.05) or CpG-Island methylation phenotype (CIMP) molecular features (AUC: 0.834$\pm$0.01 vs. 0.787$\pm$0.03, paired t-test, p$<$0.05). A combination of CIMP and SNP models further improved classification accuracy (AUC: 0.847$\pm$0.01 vs. 0.787$\pm$0.03, paired t-test, p$=$0.01). Our BP-CNN approach has the potential to provide insight into the biological cues that influence cancer histopathological imaging phenotypes and to improve the accuracy of deep-learning-based methods for determining cancer subtypes from histopathological imaging data.
Patient-level Microsatellite Stability Assessment from Whole Slide Images By Combining Momentum Contrast Learning and Group Patch Embeddings
Aug 22, 2022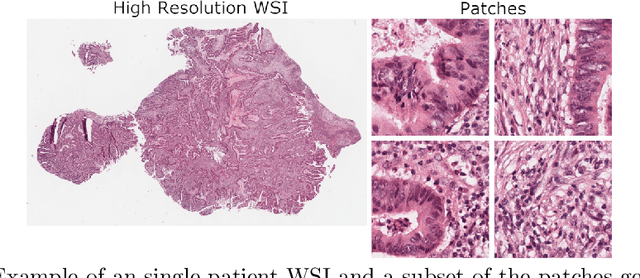
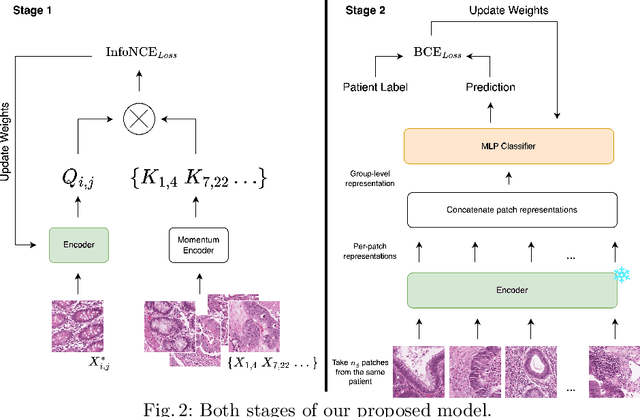
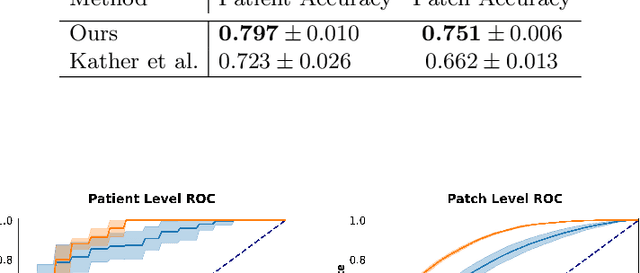
Abstract:Assessing microsatellite stability status of a patient's colorectal cancer is crucial in personalizing treatment regime. Recently, convolutional-neural-networks (CNN) combined with transfer-learning approaches were proposed to circumvent traditional laboratory testing for determining microsatellite status from hematoxylin and eosin stained biopsy whole slide images (WSI). However, the high resolution of WSI practically prevent direct classification of the entire WSI. Current approaches bypass the WSI high resolution by first classifying small patches extracted from the WSI, and then aggregating patch-level classification logits to deduce the patient-level status. Such approaches limit the capacity to capture important information which resides at the high resolution WSI data. We introduce an effective approach to leverage WSI high resolution information by momentum contrastive learning of patch embeddings along with training a patient-level classifier on groups of those embeddings. Our approach achieves up to 7.4\% better accuracy compared to the straightforward patch-level classification and patient level aggregation approach with a higher stability (AUC, $0.91 \pm 0.01$ vs. $0.85 \pm 0.04$, p-value$<0.01$). Our code can be found at https://github.com/TechnionComputationalMRILab/colorectal_cancer_ai.
WNTRAC: Artificial Intelligence Assisted Tracking of Non-pharmaceutical Interventions Implemented Worldwide for COVID-19
Sep 16, 2020



Abstract:The Coronavirus disease 2019 (COVID-19) global pandemic has transformed almost every facet of human society throughout the world. Against an emerging, highly transmissible disease with no definitive treatment or vaccine, governments worldwide have implemented non-pharmaceutical intervention (NPI) to slow the spread of the virus. Examples of such interventions include community actions (e.g. school closures, restrictions on mass gatherings), individual actions (e.g. mask wearing, self-quarantine), and environmental actions (e.g. public facility cleaning). We present the Worldwide Non-pharmaceutical Interventions Tracker for COVID-19 (WNTRAC), a comprehensive dataset consisting of over 6,000 NPIs implemented worldwide since the start of the pandemic. WNTRAC covers NPIs implemented across 261 countries and territories, and classifies NPI measures into a taxonomy of sixteen NPI types. NPI measures are automatically extracted daily from Wikipedia articles using natural language processing techniques and manually validated to ensure accuracy and veracity. We hope that the dataset is valuable for policymakers, public health leaders, and researchers in modeling and analysis efforts for controlling the spread of COVID-19.
Multi-Target Multiple Instance Learning for Hyperspectral Target Detection
Sep 07, 2019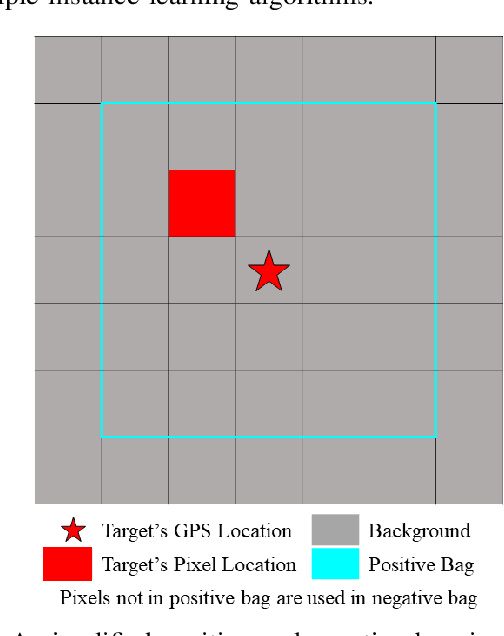
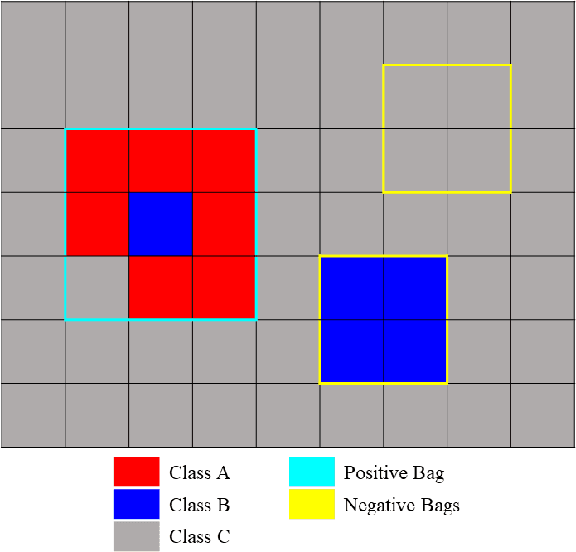
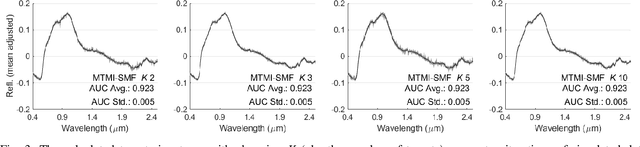
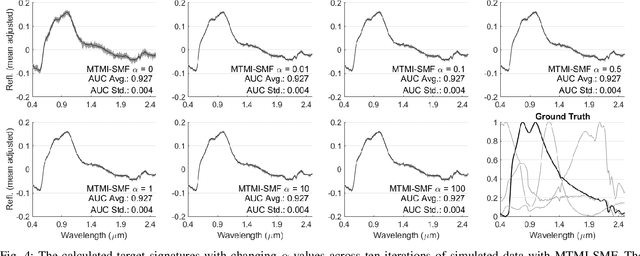
Abstract:In remote sensing, it is often difficult to acquire or collect a large dataset that is accurately labeled. This difficulty is often due to several issues including but not limited to the study site's spatial area and accessibility, errors in global positioning system (GPS), and mixed pixels caused by an image's spatial resolution. An approach, with two variations, is proposed that estimates multiple target signatures from mixed training samples with imprecise labels: Multi-Target Multiple Instance Adaptive Cosine Estimator (Multi-Target MI-ACE) and Multi-Target Multiple Instance Spectral Match Filter (Multi-Target MI-SMF). The proposed methods address the problems above by directly considering the multiple-instance, imprecisely labeled dataset and learns a dictionary of target signatures that optimizes detection using the Adaptive Cosine Estimator (ACE) and Spectral Match Filter (SMF) against a background. The algorithms have two primary steps, initialization and optimization. The initialization process determines diverse target representatives, while the optimization process simultaneously updates the target representatives to maximize detection while learning the number of optimal signatures to describe the target class. Three designed experiments were done to test the proposed algorithms: a simulated hyperspectral dataset, the MUUFL Gulfport hyperspectral dataset collected over the University of Southern Mississippi-Gulfpark Campus, and the AVIRIS hyperspectral dataset collected over Santa Barbara County, California. Both simulated and real hyperspectral target detection experiments show the proposed algorithms are effective at learning target signatures and performing target detection.
Investigation of Initialization Strategies for the Multiple Instance Adaptive Cosine Estimator
Apr 30, 2019Abstract:Sensors which use electromagnetic induction (EMI) to excite a response in conducting bodies have long been investigated for subsurface explosive hazard detection. In particular, EMI sensors have been used to discriminate between different types of objects, and to detect objects with low metal content. One successful, previously investigated approach is the Multiple Instance Adaptive Cosine Estimator (MI-ACE). In this paper, a number of new initialization techniques for MI-ACE are proposed and evaluated using their respective performance and speed. The cross validated learned signatures, as well as learned background statistics, are used with Adaptive Cosine Estimator (ACE) to generate confidence maps, which are clustered into alarms. Alarms are scored against a ground truth and the initialization approaches are compared.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge