Sivan Ravid
MAMMAL -- Molecular Aligned Multi-Modal Architecture and Language
Oct 28, 2024Abstract:Drug discovery typically consists of multiple steps, including identifying a target protein key to a disease's etiology, validating that interacting with this target could prevent symptoms or cure the disease, discovering a small molecule or biologic therapeutic to interact with it, and optimizing the candidate molecule through a complex landscape of required properties. Drug discovery related tasks often involve prediction and generation while considering multiple entities that potentially interact, which poses a challenge for typical AI models. For this purpose we present MAMMAL - Molecular Aligned Multi-Modal Architecture and Language - a method that we applied to create a versatile multi-task foundation model ibm/biomed.omics.bl.sm.ma-ted-458m that learns from large-scale biological datasets (2 billion samples) across diverse modalities, including proteins, small molecules, and genes. We introduce a prompt syntax that supports a wide range of classification, regression, and generation tasks. It allows combining different modalities and entity types as inputs and/or outputs. Our model handles combinations of tokens and scalars and enables the generation of small molecules and proteins, property prediction, and transcriptomic lab test predictions. We evaluated the model on 11 diverse downstream tasks spanning different steps within a typical drug discovery pipeline, where it reaches new SOTA in 9 tasks and is comparable to SOTA in 2 tasks. This performance is achieved while using a unified architecture serving all tasks, in contrast to the original SOTA performance achieved using tailored architectures. The model code and pretrained weights are publicly available at https://github.com/BiomedSciAI/biomed-multi-alignment and https://huggingface.co/ibm/biomed.omics.bl.sm.ma-ted-458m.
A large dataset curation and benchmark for drug target interaction
Jan 30, 2024Abstract:Bioactivity data plays a key role in drug discovery and repurposing. The resource-demanding nature of \textit{in vitro} and \textit{in vivo} experiments, as well as the recent advances in data-driven computational biochemistry research, highlight the importance of \textit{in silico} drug target interaction (DTI) prediction approaches. While numerous large public bioactivity data sources exist, research in the field could benefit from better standardization of existing data resources. At present, different research works that share similar goals are often difficult to compare properly because of different choices of data sources and train/validation/test split strategies. Additionally, many works are based on small data subsets, leading to results and insights of possible limited validity. In this paper we propose a way to standardize and represent efficiently a very large dataset curated from multiple public sources, split the data into train, validation and test sets based on different meaningful strategies, and provide a concrete evaluation protocol to accomplish a benchmark. We analyze the proposed data curation, prove its usefulness and validate the proposed benchmark through experimental studies based on an existing neural network model.
An Evaluation Toolkit to Guide Model Selection and Cohort Definition in Causal Inference
Jun 02, 2019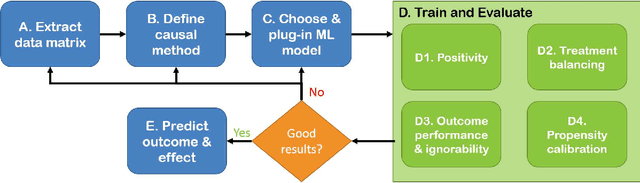

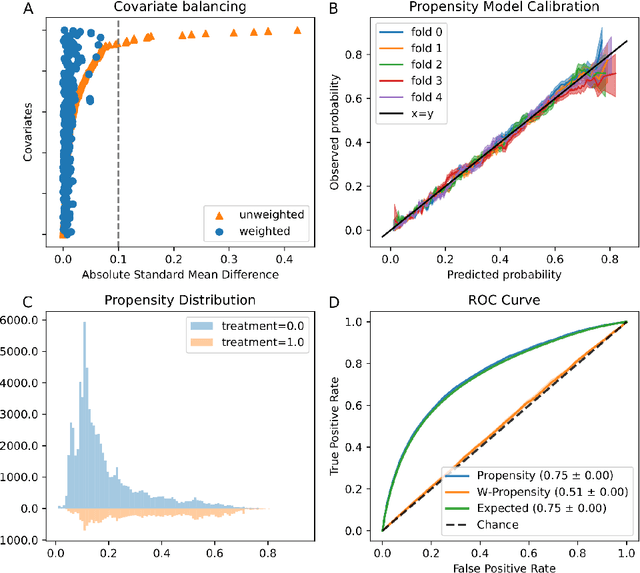
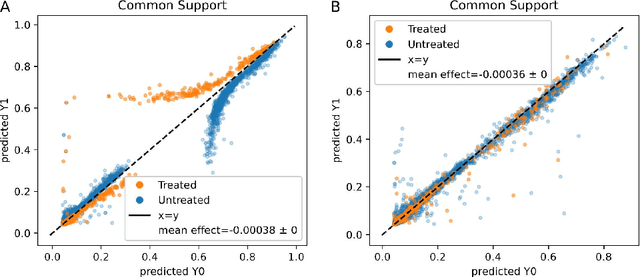
Abstract:Real world observational data, together with causal inference, allow the estimation of causal effects when randomized controlled trials are not available. To be accepted into practice, such predictive models must be validated for the dataset at hand, and thus require a comprehensive evaluation toolkit, as introduced here. Since effect estimation cannot be evaluated directly, we turn to evaluating the various observable properties of causal inference, namely the observed outcome and treatment assignment. We developed a toolkit that expands established machine learning evaluation methods and adds several causal-specific ones. Evaluations can be applied in cross-validation, in a train-test scheme, or on the training data. Multiple causal inference methods are implemented within the toolkit in a way that allows modular use of the underlying machine learning models. Thus, the toolkit is agnostic to the machine learning model that is used. We showcase our approach using a rheumatoid arthritis cohort (consisting of about 120K patients) extracted from the IBM MarketScan(R) Research Database. We introduce an iterative pipeline of data definition, model definition, and model evaluation. Using this pipeline, we demonstrate how each of the evaluation components helps drive model selection and refinement of data extraction criteria in a way that provides more reproducible results and ensures that the causal question is answerable with available data. Furthermore, we show how the evaluation toolkit can be used to ensure that performance is maintained when applied to subsets of the data, thus allowing exploration of questions that move towards personalized medicine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge