Vadim Ratner
MAMMAL -- Molecular Aligned Multi-Modal Architecture and Language
Oct 28, 2024Abstract:Drug discovery typically consists of multiple steps, including identifying a target protein key to a disease's etiology, validating that interacting with this target could prevent symptoms or cure the disease, discovering a small molecule or biologic therapeutic to interact with it, and optimizing the candidate molecule through a complex landscape of required properties. Drug discovery related tasks often involve prediction and generation while considering multiple entities that potentially interact, which poses a challenge for typical AI models. For this purpose we present MAMMAL - Molecular Aligned Multi-Modal Architecture and Language - a method that we applied to create a versatile multi-task foundation model ibm/biomed.omics.bl.sm.ma-ted-458m that learns from large-scale biological datasets (2 billion samples) across diverse modalities, including proteins, small molecules, and genes. We introduce a prompt syntax that supports a wide range of classification, regression, and generation tasks. It allows combining different modalities and entity types as inputs and/or outputs. Our model handles combinations of tokens and scalars and enables the generation of small molecules and proteins, property prediction, and transcriptomic lab test predictions. We evaluated the model on 11 diverse downstream tasks spanning different steps within a typical drug discovery pipeline, where it reaches new SOTA in 9 tasks and is comparable to SOTA in 2 tasks. This performance is achieved while using a unified architecture serving all tasks, in contrast to the original SOTA performance achieved using tailored architectures. The model code and pretrained weights are publicly available at https://github.com/BiomedSciAI/biomed-multi-alignment and https://huggingface.co/ibm/biomed.omics.bl.sm.ma-ted-458m.
A large dataset curation and benchmark for drug target interaction
Jan 30, 2024Abstract:Bioactivity data plays a key role in drug discovery and repurposing. The resource-demanding nature of \textit{in vitro} and \textit{in vivo} experiments, as well as the recent advances in data-driven computational biochemistry research, highlight the importance of \textit{in silico} drug target interaction (DTI) prediction approaches. While numerous large public bioactivity data sources exist, research in the field could benefit from better standardization of existing data resources. At present, different research works that share similar goals are often difficult to compare properly because of different choices of data sources and train/validation/test split strategies. Additionally, many works are based on small data subsets, leading to results and insights of possible limited validity. In this paper we propose a way to standardize and represent efficiently a very large dataset curated from multiple public sources, split the data into train, validation and test sets based on different meaningful strategies, and provide a concrete evaluation protocol to accomplish a benchmark. We analyze the proposed data curation, prove its usefulness and validate the proposed benchmark through experimental studies based on an existing neural network model.
Regularized adversarial examples for model interpretability
Nov 21, 2018


Abstract:As machine learning algorithms continue to improve, there is an increasing need for explaining why a model produces a certain prediction for a certain input. In recent years, several methods for model interpretability have been developed, aiming to provide explanation of which subset regions of the model input is the main reason for the model prediction. In parallel, a significant research community effort is occurring in recent years for developing adversarial example generation methods for fooling models, while not altering the true label of the input,as it would have been classified by a human annotator. In this paper, we bridge the gap between adversarial example generation and model interpretability, and introduce a modification to the adversarial example generation process which encourages better interpretability. We analyze the proposed method on a public medical imaging dataset, both quantitatively and qualitatively, and show that it significantly outperforms the leading known alternative method. Our suggested method is simple to implement, and can be easily plugged into most common adversarial example generation frameworks. Additionally, we propose an explanation quality metric - $APE$ - "Adversarial Perturbative Explanation", which measures how well an explanation describes model decisions.
Learning multiple non-mutually-exclusive tasks for improved classification of inherently ordered labels
May 30, 2018

Abstract:Medical image classification involves thresholding of labels that represent malignancy risk levels. Usually, a task defines a single threshold, and when developing computer-aided diagnosis tools, a single network is trained per such threshold, e.g. as screening out healthy (very low risk) patients to leave possibly sick ones for further analysis (low threshold), or trying to find malignant cases among those marked as non-risk by the radiologist ("second reading", high threshold). We propose a way to rephrase the classification problem in a manner that yields several problems (corresponding to different thresholds) to be solved simultaneously. This allows the use of Multiple Task Learning (MTL) methods, significantly improving the performance of the original classifier, by facilitating effective extraction of information from existing data.
AdapterNet - learning input transformation for domain adaptation
May 29, 2018

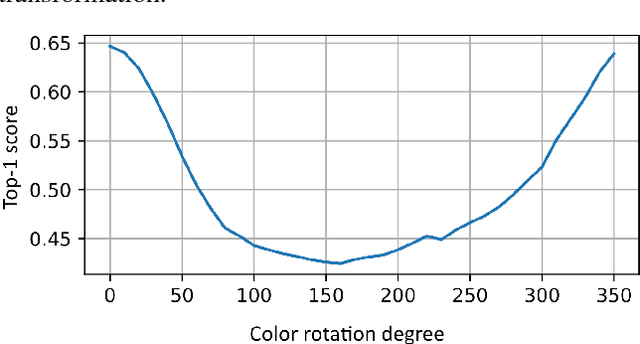

Abstract:Deep neural networks have demonstrated impressive performance in various machine learning tasks. However, they are notoriously sensitive to changes in data distribution. Often, even a slight change in the distribution can lead to drastic performance reduction. Artificially augmenting the data may help to some extent, but in most cases, fails to achieve model invariance to the data distribution. Some examples where this sub-class of domain adaptation can be valuable are various imaging modalities such as thermal imaging, X-ray, ultrasound, and MRI, where changes in acquisition parameters or acquisition device manufacturer will result in different representation of the same input. Our work shows that standard finetuning fails to adapt the model in certain important cases. We propose a novel method of adapting to a new data source, and demonstrate near perfect adaptation on a customized ImageNet benchmark.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge