Moshiko Raboh
MAMMAL -- Molecular Aligned Multi-Modal Architecture and Language
Oct 28, 2024Abstract:Drug discovery typically consists of multiple steps, including identifying a target protein key to a disease's etiology, validating that interacting with this target could prevent symptoms or cure the disease, discovering a small molecule or biologic therapeutic to interact with it, and optimizing the candidate molecule through a complex landscape of required properties. Drug discovery related tasks often involve prediction and generation while considering multiple entities that potentially interact, which poses a challenge for typical AI models. For this purpose we present MAMMAL - Molecular Aligned Multi-Modal Architecture and Language - a method that we applied to create a versatile multi-task foundation model ibm/biomed.omics.bl.sm.ma-ted-458m that learns from large-scale biological datasets (2 billion samples) across diverse modalities, including proteins, small molecules, and genes. We introduce a prompt syntax that supports a wide range of classification, regression, and generation tasks. It allows combining different modalities and entity types as inputs and/or outputs. Our model handles combinations of tokens and scalars and enables the generation of small molecules and proteins, property prediction, and transcriptomic lab test predictions. We evaluated the model on 11 diverse downstream tasks spanning different steps within a typical drug discovery pipeline, where it reaches new SOTA in 9 tasks and is comparable to SOTA in 2 tasks. This performance is achieved while using a unified architecture serving all tasks, in contrast to the original SOTA performance achieved using tailored architectures. The model code and pretrained weights are publicly available at https://github.com/BiomedSciAI/biomed-multi-alignment and https://huggingface.co/ibm/biomed.omics.bl.sm.ma-ted-458m.
Differentiable Scene Graphs
Mar 26, 2019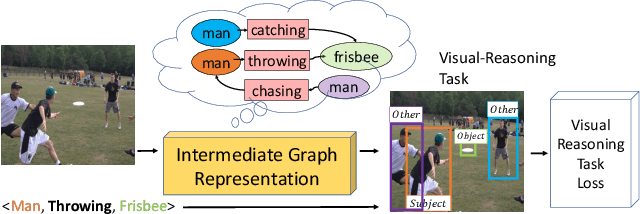
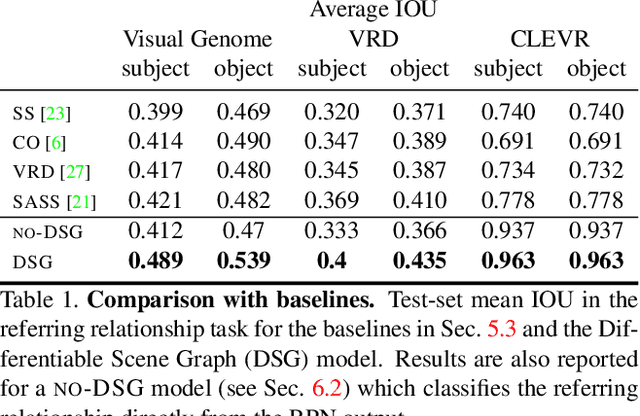
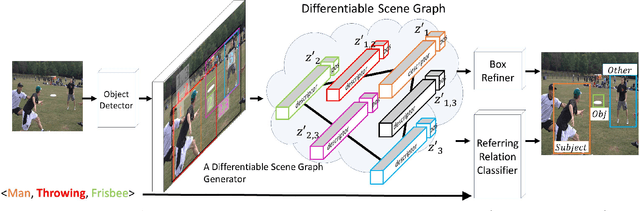
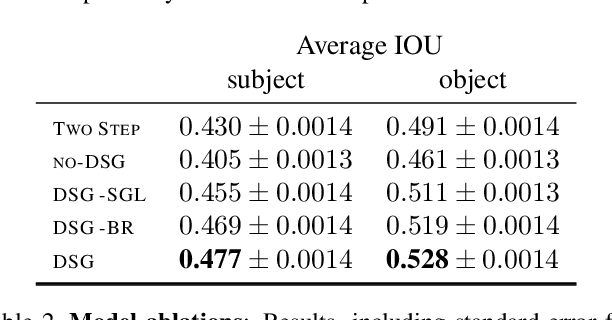
Abstract:Understanding the semantics of complex visual scenes involves perception of entities and reasoning about their relations. Scene graphs provide a natural representation for these tasks, by assigning labels to both entities (nodes) and relations (edges). However, scene graphs are not commonly used as intermediate components in visual reasoning systems, for two complementary reasons. First, training models to map images to scene graphs requires prohibitive manual annotation, and results in graphs that often do not match the needs of a downstream visual reasoning application. Second, using these discrete graphs as an intermediate latent representation results in a non-differentiable function that is difficult to optimize. Here we propose Differentiable Scene Graphs (DSGs), an image representation that is amenable to differentiable end-to-end optimization, and requires supervision only from the downstream tasks. DSGs provide a dense representation for all regions and pairs of regions, investing model capacity on the important aspects of the image. We describe a multi-task objective function that allows us to learn this representation from indirect supervision only, provided by the downstream task. We evaluate our model on the challenging task of identifying referring relationships, and show that training DSGs using our multi-task objective leads to new state-of-the-art performance.
Mapping Images to Scene Graphs with Permutation-Invariant Structured Prediction
Nov 01, 2018
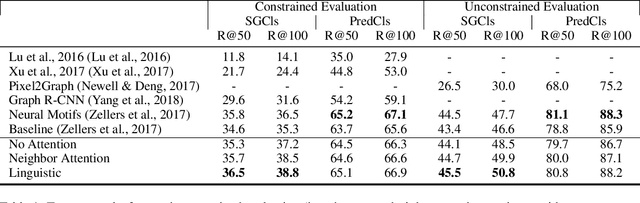
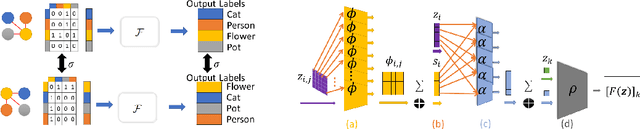

Abstract:Machine understanding of complex images is a key goal of artificial intelligence. One challenge underlying this task is that visual scenes contain multiple inter-related objects, and that global context plays an important role in interpreting the scene. A natural modeling framework for capturing such effects is structured prediction, which optimizes over complex labels, while modeling within-label interactions. However, it is unclear what principles should guide the design of a structured prediction model that utilizes the power of deep learning components. Here we propose a design principle for such architectures that follows from a natural requirement of permutation invariance. We prove a necessary and sufficient characterization for architectures that follow this invariance, and discuss its implication on model design. Finally, we show that the resulting model achieves new state of the art results on the Visual Genome scene graph labeling benchmark, outperforming all recent approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge