Liam Hazan
MAMMAL -- Molecular Aligned Multi-Modal Architecture and Language
Oct 28, 2024Abstract:Drug discovery typically consists of multiple steps, including identifying a target protein key to a disease's etiology, validating that interacting with this target could prevent symptoms or cure the disease, discovering a small molecule or biologic therapeutic to interact with it, and optimizing the candidate molecule through a complex landscape of required properties. Drug discovery related tasks often involve prediction and generation while considering multiple entities that potentially interact, which poses a challenge for typical AI models. For this purpose we present MAMMAL - Molecular Aligned Multi-Modal Architecture and Language - a method that we applied to create a versatile multi-task foundation model ibm/biomed.omics.bl.sm.ma-ted-458m that learns from large-scale biological datasets (2 billion samples) across diverse modalities, including proteins, small molecules, and genes. We introduce a prompt syntax that supports a wide range of classification, regression, and generation tasks. It allows combining different modalities and entity types as inputs and/or outputs. Our model handles combinations of tokens and scalars and enables the generation of small molecules and proteins, property prediction, and transcriptomic lab test predictions. We evaluated the model on 11 diverse downstream tasks spanning different steps within a typical drug discovery pipeline, where it reaches new SOTA in 9 tasks and is comparable to SOTA in 2 tasks. This performance is achieved while using a unified architecture serving all tasks, in contrast to the original SOTA performance achieved using tailored architectures. The model code and pretrained weights are publicly available at https://github.com/BiomedSciAI/biomed-multi-alignment and https://huggingface.co/ibm/biomed.omics.bl.sm.ma-ted-458m.
Leveraging Prompt-Learning for Structured Information Extraction from Crohn's Disease Radiology Reports in a Low-Resource Language
May 02, 2024Abstract:Automatic conversion of free-text radiology reports into structured data using Natural Language Processing (NLP) techniques is crucial for analyzing diseases on a large scale. While effective for tasks in widely spoken languages like English, generative large language models (LLMs) typically underperform with less common languages and can pose potential risks to patient privacy. Fine-tuning local NLP models is hindered by the skewed nature of real-world medical datasets, where rare findings represent a significant data imbalance. We introduce SMP-BERT, a novel prompt learning method that leverages the structured nature of reports to overcome these challenges. In our studies involving a substantial collection of Crohn's disease radiology reports in Hebrew (over 8,000 patients and 10,000 reports), SMP-BERT greatly surpassed traditional fine-tuning methods in performance, notably in detecting infrequent conditions (AUC: 0.99 vs 0.94, F1: 0.84 vs 0.34). SMP-BERT empowers more accurate AI diagnostics available for low-resource languages.
A large dataset curation and benchmark for drug target interaction
Jan 30, 2024Abstract:Bioactivity data plays a key role in drug discovery and repurposing. The resource-demanding nature of \textit{in vitro} and \textit{in vivo} experiments, as well as the recent advances in data-driven computational biochemistry research, highlight the importance of \textit{in silico} drug target interaction (DTI) prediction approaches. While numerous large public bioactivity data sources exist, research in the field could benefit from better standardization of existing data resources. At present, different research works that share similar goals are often difficult to compare properly because of different choices of data sources and train/validation/test split strategies. Additionally, many works are based on small data subsets, leading to results and insights of possible limited validity. In this paper we propose a way to standardize and represent efficiently a very large dataset curated from multiple public sources, split the data into train, validation and test sets based on different meaningful strategies, and provide a concrete evaluation protocol to accomplish a benchmark. We analyze the proposed data curation, prove its usefulness and validate the proposed benchmark through experimental studies based on an existing neural network model.
BLOOM: A 176B-Parameter Open-Access Multilingual Language Model
Nov 09, 2022Abstract:Large language models (LLMs) have been shown to be able to perform new tasks based on a few demonstrations or natural language instructions. While these capabilities have led to widespread adoption, most LLMs are developed by resource-rich organizations and are frequently kept from the public. As a step towards democratizing this powerful technology, we present BLOOM, a 176B-parameter open-access language model designed and built thanks to a collaboration of hundreds of researchers. BLOOM is a decoder-only Transformer language model that was trained on the ROOTS corpus, a dataset comprising hundreds of sources in 46 natural and 13 programming languages (59 in total). We find that BLOOM achieves competitive performance on a wide variety of benchmarks, with stronger results after undergoing multitask prompted finetuning. To facilitate future research and applications using LLMs, we publicly release our models and code under the Responsible AI License.
Length Learning for Planar Euclidean Curves
Feb 03, 2021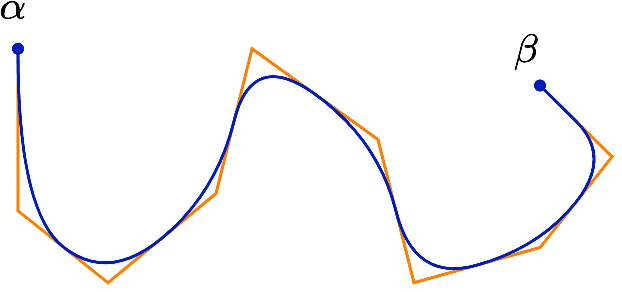
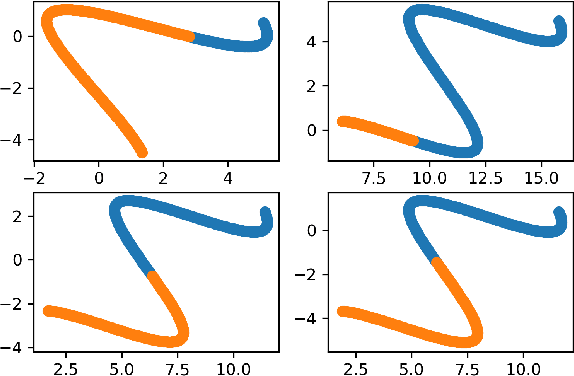
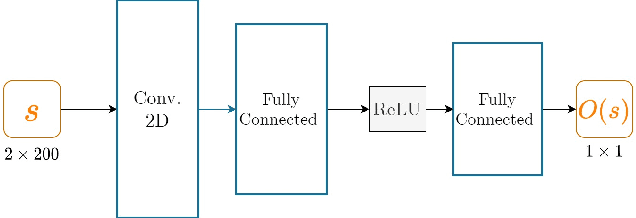
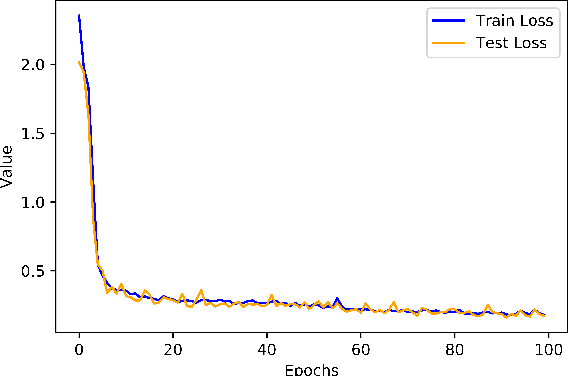
Abstract:In this work, we used deep neural networks (DNNs) to solve a fundamental problem in differential geometry. One can find many closed-form expressions for calculating curvature, length, and other geometric properties in the literature. As we know these concepts, we are highly motivated to reconstruct them by using deep neural networks. In this framework, our goal is to learn geometric properties from examples. The simplest geometric object is a curve. Therefore, this work focuses on learning the length of planar sampled curves created by a sine waves dataset. For this reason, the fundamental length axioms were reconstructed using a supervised learning approach. Following these axioms a simplified DNN model, we call ArcLengthNet, was established. The robustness to additive noise and discretization errors were tested.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge