Huai Chen
The KiTS21 Challenge: Automatic segmentation of kidneys, renal tumors, and renal cysts in corticomedullary-phase CT
Jul 05, 2023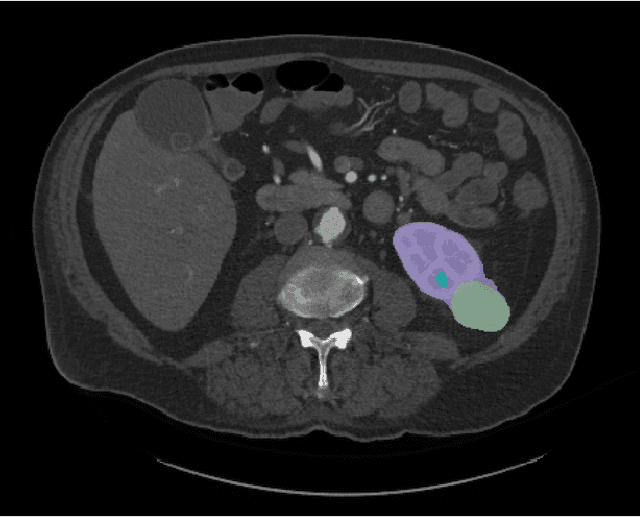
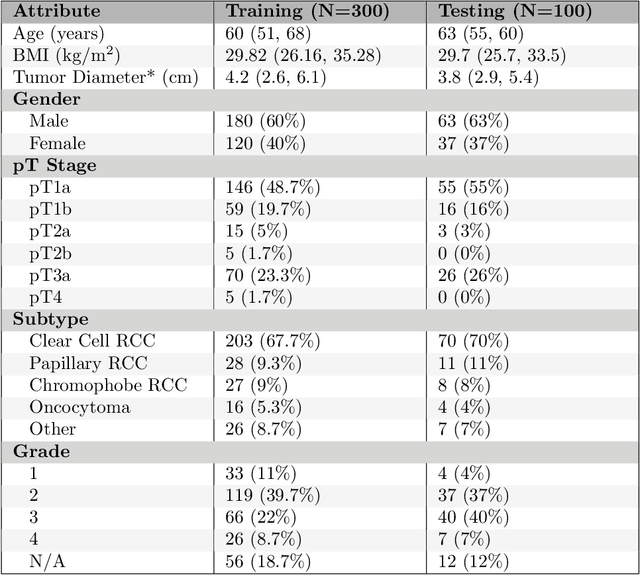
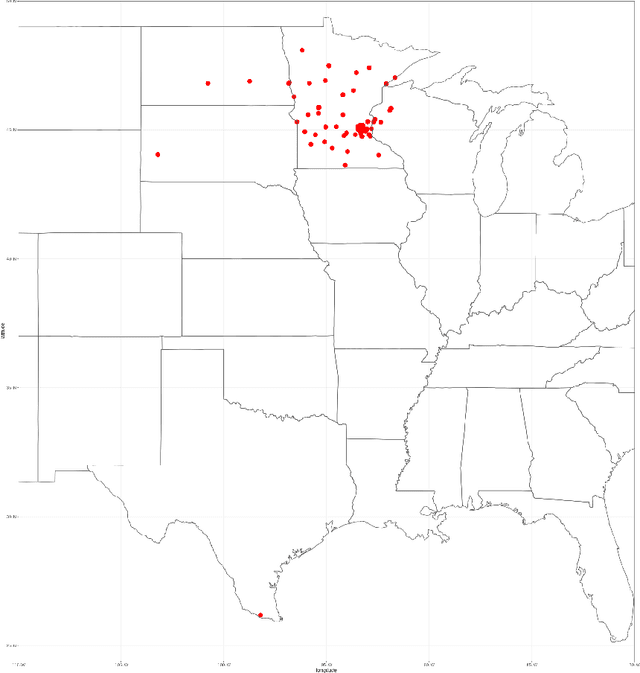
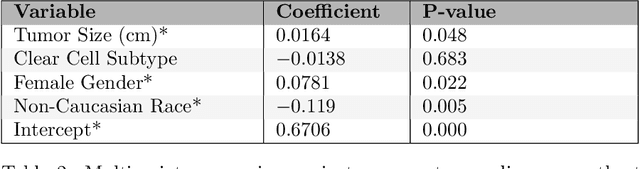
Abstract:This paper presents the challenge report for the 2021 Kidney and Kidney Tumor Segmentation Challenge (KiTS21) held in conjunction with the 2021 international conference on Medical Image Computing and Computer Assisted Interventions (MICCAI). KiTS21 is a sequel to its first edition in 2019, and it features a variety of innovations in how the challenge was designed, in addition to a larger dataset. A novel annotation method was used to collect three separate annotations for each region of interest, and these annotations were performed in a fully transparent setting using a web-based annotation tool. Further, the KiTS21 test set was collected from an outside institution, challenging participants to develop methods that generalize well to new populations. Nonetheless, the top-performing teams achieved a significant improvement over the state of the art set in 2019, and this performance is shown to inch ever closer to human-level performance. An in-depth meta-analysis is presented describing which methods were used and how they faired on the leaderboard, as well as the characteristics of which cases generally saw good performance, and which did not. Overall KiTS21 facilitated a significant advancement in the state of the art in kidney tumor segmentation, and provides useful insights that are applicable to the field of semantic segmentation as a whole.
3D Vessel Segmentation with Limited Guidance of 2D Structure-agnostic Vessel Annotations
Feb 07, 2023



Abstract:Delineating 3D blood vessels is essential for clinical diagnosis and treatment, however, is challenging due to complex structure variations and varied imaging conditions. Supervised deep learning has demonstrated its superior capacity in automatic 3D vessel segmentation. However, the reliance on expensive 3D manual annotations and limited capacity for annotation reuse hinder the clinical applications of supervised models. To avoid the repetitive and laborious annotating and make full use of existing vascular annotations, this paper proposes a novel 3D shape-guided local discrimination model for 3D vascular segmentation under limited guidance from public 2D vessel annotations. The primary hypothesis is that 3D vessels are composed of semantically similar voxels and exhibit tree-shaped morphology. Accordingly, the 3D region discrimination loss is firstly proposed to learn the discriminative representation measuring voxel-wise similarities and cluster semantically consistent voxels to form the candidate 3D vascular segmentation in unlabeled images; secondly, based on the similarity of the tree-shaped morphology between 2D and 3D vessels, the Crop-and-Overlap strategy is presented to generate reference masks from 2D structure-agnostic vessel annotations, which are fit for varied vascular structures, and the adversarial loss is introduced to guide the tree-shaped morphology of 3D vessels; thirdly, the temporal consistency loss is proposed to foster the training stability and keep the model updated smoothly. To further enhance the model's robustness and reliability, the orientation-invariant CNN module and Reliability-Refinement algorithm are presented. Experimental results from the public 3D cerebrovascular and 3D arterial tree datasets demonstrate that our model achieves comparable effectiveness against nine supervised models.
Unsupervised Local Discrimination for Medical Images
Aug 21, 2021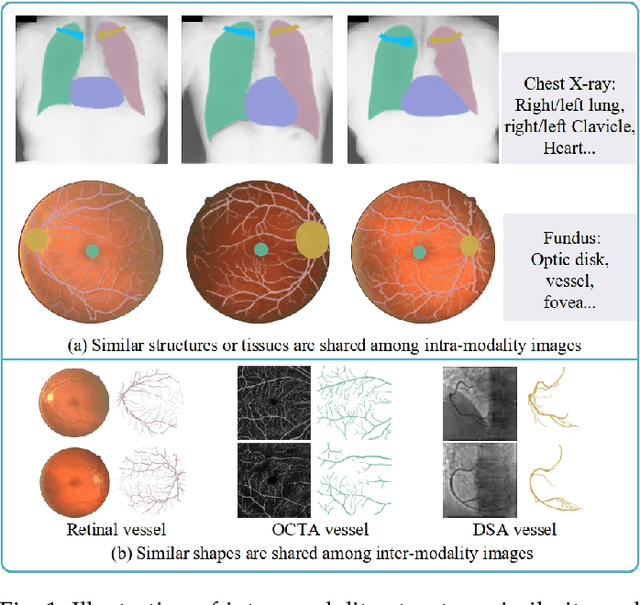
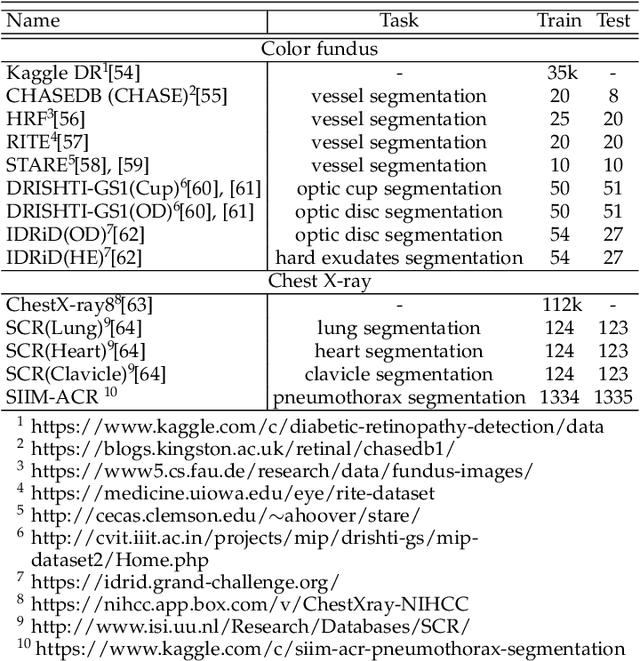
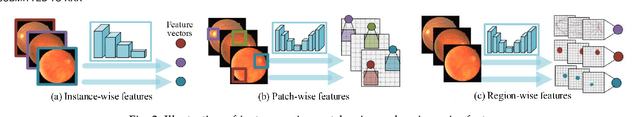
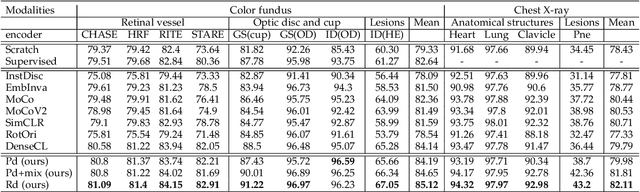
Abstract:Contrastive representation learning is an effective unsupervised method to alleviate the demand for expensive annotated data in medical image processing. Recent work mainly based on instance-wise discrimination to learn global features, while neglect local details, which limit their application in processing tiny anatomical structures, tissues and lesions. Therefore, we aim to propose a universal local discrmination framework to learn local discriminative features to effectively initialize medical models, meanwhile, we systematacially investigate its practical medical applications. Specifically, based on the common property of intra-modality structure similarity, i.e. similar structures are shared among the same modality images, a systematic local feature learning framework is proposed. Instead of making instance-wise comparisons based on global embedding, our method makes pixel-wise embedding and focuses on measuring similarity among patches and regions. The finer contrastive rule makes the learnt representation more generalized for segmentation tasks and outperform extensive state-of-the-art methods by wining 11 out of all 12 downstream tasks in color fundus and chest X-ray. Furthermore, based on the property of inter-modality shape similarity, i.e. structures may share similar shape although in different medical modalities, we joint across-modality shape prior into region discrimination to realize unsupervised segmentation. It shows the feaibility of segmenting target only based on shape description from other modalities and inner pattern similarity provided by region discrimination. Finally, we enhance the center-sensitive ability of patch discrimination by introducing center-sensitive averaging to realize one-shot landmark localization, this is an effective application for patch discrimination.
COVID-MTL: Multitask Learning with Shift3D and Random-weighted Loss for Automated Diagnosis and Severity Assessment of COVID-19
Dec 31, 2020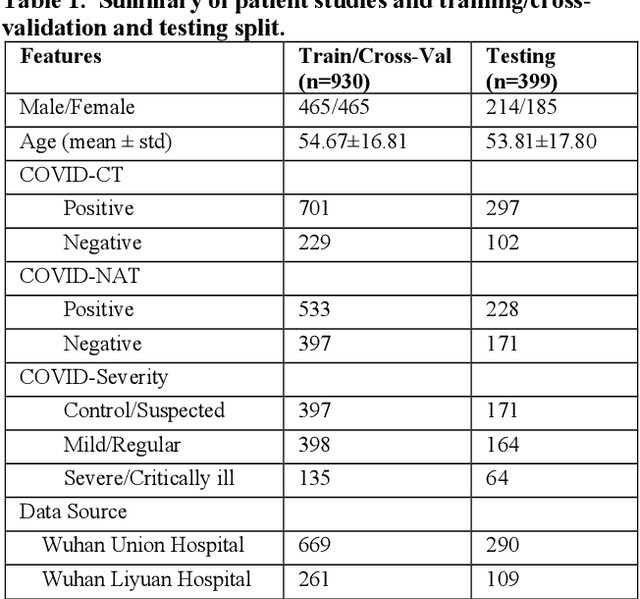
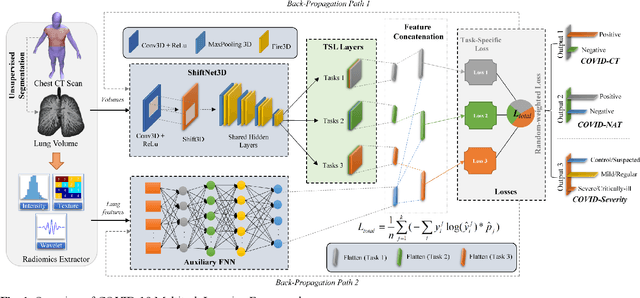
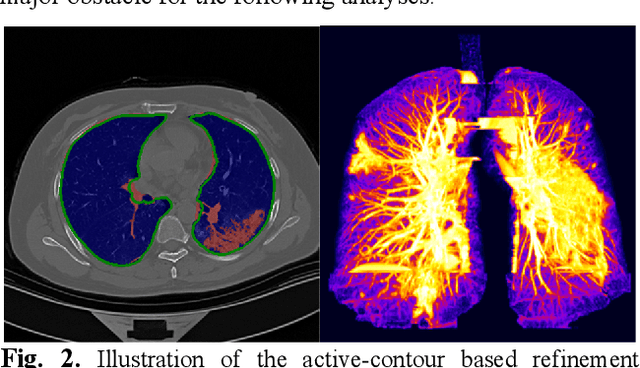
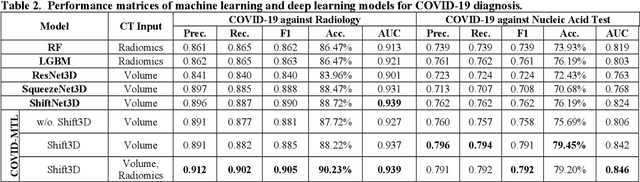
Abstract:There is an urgent need for automated methods to assist accurate and effective assessment of COVID-19. Radiology and nucleic acid test (NAT) are complementary COVID-19 diagnosis methods. In this paper, we present an end-to-end multitask learning (MTL) framework (COVID-MTL) that is capable of automated and simultaneous detection (against both radiology and NAT) and severity assessment of COVID-19. COVID-MTL learns different COVID-19 tasks in parallel through our novel random-weighted loss function, which assigns learning weights under Dirichlet distribution to prevent task dominance; our new 3D real-time augmentation algorithm (Shift3D) introduces space variances for 3D CNN components by shifting low-level feature representations of volumetric inputs in three dimensions; thereby, the MTL framework is able to accelerate convergence and improve joint learning performance compared to single-task models. By only using chest CT scans, COVID-MTL was trained on 930 CT scans and tested on separate 399 cases. COVID-MTL achieved AUCs of 0.939 and 0.846, and accuracies of 90.23% and 79.20% for detection of COVID-19 against radiology and NAT, respectively, which outperformed the state-of-the-art models. Meanwhile, COVID-MTL yielded AUC of 0.800 $\pm$ 0.020 and 0.813 $\pm$ 0.021 (with transfer learning) for classifying control/suspected, mild/regular, and severe/critically-ill cases. To decipher the recognition mechanism, we also identified high-throughput lung features that were significantly related (P < 0.001) to the positivity and severity of COVID-19.
Unsupervised Learning of Local Discriminative Representation for Medical Images
Dec 17, 2020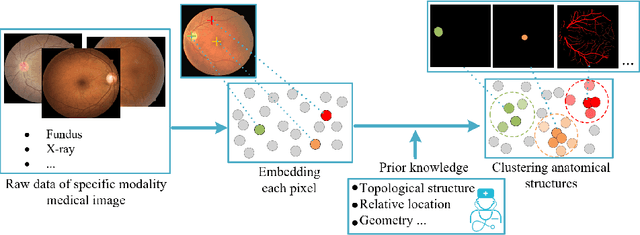
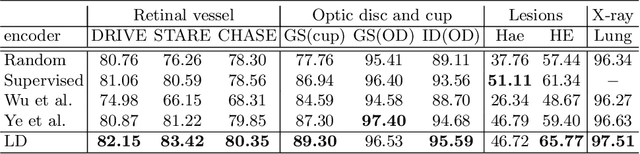
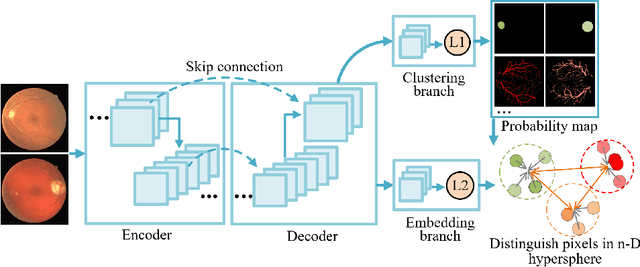
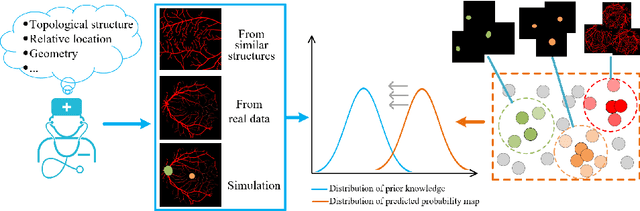
Abstract:Local discriminative representation is needed in many medical image analysis tasks such as identifying sub-types of lesion or segmenting detailed components of anatomical structures by measuring similarity of local image regions. However, the commonly applied supervised representation learning methods require a large amount of annotated data, and unsupervised discriminative representation learning distinguishes different images by learning a global feature. In order to avoid the limitations of these two methods and be suitable for localized medical image analysis tasks, we introduce local discrimination into unsupervised representation learning in this work. The model contains two branches: one is an embedding branch which learns an embedding function to disperse dissimilar pixels over a low-dimensional hypersphere; and the other is a clustering branch which learns a clustering function to classify similar pixels into the same cluster. These two branches are trained simultaneously in a mutually beneficial pattern, and the learnt local discriminative representations are able to well measure the similarity of local image regions. These representations can be transferred to enhance various downstream tasks. Meanwhile, they can also be applied to cluster anatomical structures from unlabeled medical images under the guidance of topological priors from simulation or other structures with similar topological characteristics. The effectiveness and usefulness of the proposed method are demonstrated by enhancing various downstream tasks and clustering anatomical structures in retinal images and chest X-ray images. The corresponding code is available at https://github.com/HuaiChen-1994/LDLearning.
MMFNet: A Multi-modality MRI Fusion Network for Segmentation of Nasopharyngeal Carcinoma
Jan 22, 2019



Abstract:Segmentation of nasopharyngeal carcinoma (NPC) from Magnetic Resonance Images (MRI) is a crucial step in NPC radiotherapy. However, manually segmenting of NPC is a time-consuming and labor-intensive task. Additionally, single-modality MRI generally cannot provide enough information for the accurate delineation of NPC. Therefore, a multi-modality MRI fusion network (MMFNet) based on three modalities of MRI (T1, T2 and contrast-enhanced T1) is proposed to complete accurate segmentation of NPC. In the MMFNet, the backbone is designed as a multi-encoder-based network, consisting of several modality-specific encoders and one single decoder. It can be used to well learn both low-level and high-level features used implicitly for NPC segmentation in each modality of MRI. A fusion block is proposed in the MMFNet to effectively fuse low-level features from multi-modality MRI. It firstly recalibrates features captured from multi-modality MRI, which will highlight informative features and regions of interest. Then, a residual fusion block is utilized to fuse weighted features before merging them with features from decoder to keep balance between high-level and low-level features. Moreover, a training strategy named self-transfer is proposed to initialize encoders for multi-encoder-based network. It can stimulate encoders to make full mining of modality-specific MRI. The proposed method can effectively make use of information in multi-modality MRI. Its effectiveness and advantages are validated by many experiments and comparisons with the related methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge