Zhongchen Zhao
The KiTS21 Challenge: Automatic segmentation of kidneys, renal tumors, and renal cysts in corticomedullary-phase CT
Jul 05, 2023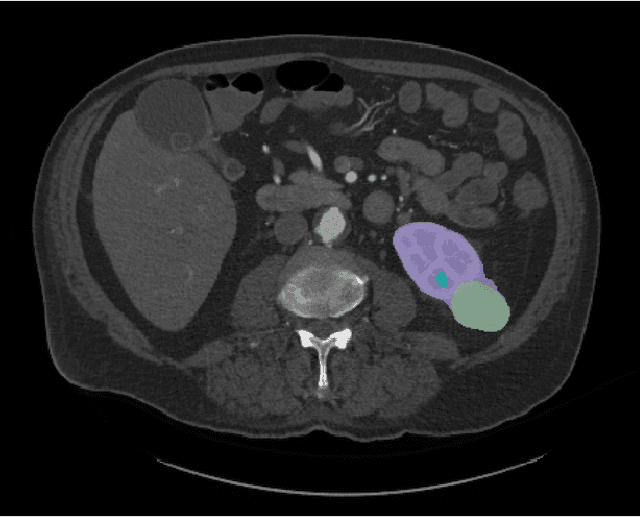
Abstract:This paper presents the challenge report for the 2021 Kidney and Kidney Tumor Segmentation Challenge (KiTS21) held in conjunction with the 2021 international conference on Medical Image Computing and Computer Assisted Interventions (MICCAI). KiTS21 is a sequel to its first edition in 2019, and it features a variety of innovations in how the challenge was designed, in addition to a larger dataset. A novel annotation method was used to collect three separate annotations for each region of interest, and these annotations were performed in a fully transparent setting using a web-based annotation tool. Further, the KiTS21 test set was collected from an outside institution, challenging participants to develop methods that generalize well to new populations. Nonetheless, the top-performing teams achieved a significant improvement over the state of the art set in 2019, and this performance is shown to inch ever closer to human-level performance. An in-depth meta-analysis is presented describing which methods were used and how they faired on the leaderboard, as well as the characteristics of which cases generally saw good performance, and which did not. Overall KiTS21 facilitated a significant advancement in the state of the art in kidney tumor segmentation, and provides useful insights that are applicable to the field of semantic segmentation as a whole.
Exploring Visual Prompts for Whole Slide Image Classification with Multiple Instance Learning
Mar 23, 2023



Abstract:Multiple instance learning (MIL) has emerged as a popular method for classifying histopathology whole slide images (WSIs). However, existing approaches typically rely on pre-trained models from large natural image datasets, such as ImageNet, to generate instance features, which can be sub-optimal due to the significant differences between natural images and histopathology images that lead to a domain shift. In this paper, we present a novel, simple yet effective method for learning domain-specific knowledge transformation from pre-trained models to histopathology images. Our approach entails using a prompt component to assist the pre-trained model in discerning differences between the pre-trained dataset and the target histopathology dataset, resulting in improved performance of MIL models. We validate our method on two publicly available datasets, Camelyon16 and TCGA-NSCLC. Extensive experimental results demonstrate the significant performance improvement of our method for different MIL models and backbones. Upon publication of this paper, we will release the source code for our method.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
A Multi-Stage Framework for the 2022 Multi-Structure Segmentation for Renal Cancer Treatment
Jul 19, 2022
Abstract:Three-dimensional (3D) kidney parsing on computed tomography angiography (CTA) images is of great clinical significance. Automatic segmentation of kidney, renal tumor, renal vein and renal artery benefits a lot on surgery-based renal cancer treatment. In this paper, we propose a new nnhra-unet network, and use a multi-stage framework which is based on it to segment the multi-structure of kidney and participate in the KiPA2022 challenge.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge