Kwang-Ting Cheng
School of Engineering, Hong Kong University of Science and Technology
A Study of Finetuning Video Transformers for Multi-view Geometry Tasks
Dec 21, 2025Abstract:This paper presents an investigation of vision transformer learning for multi-view geometry tasks, such as optical flow estimation, by fine-tuning video foundation models. Unlike previous methods that involve custom architectural designs and task-specific pretraining, our research finds that general-purpose models pretrained on videos can be readily transferred to multi-view problems with minimal adaptation. The core insight is that general-purpose attention between patches learns temporal and spatial information for geometric reasoning. We demonstrate that appending a linear decoder to the Transformer backbone produces satisfactory results, and iterative refinement can further elevate performance to stateof-the-art levels. This conceptually simple approach achieves top cross-dataset generalization results for optical flow estimation with end-point error (EPE) of 0.69, 1.78, and 3.15 on the Sintel clean, Sintel final, and KITTI datasets, respectively. Our method additionally establishes a new record on the online test benchmark with EPE values of 0.79, 1.88, and F1 value of 3.79. Applications to 3D depth estimation and stereo matching also show strong performance, illustrating the versatility of video-pretrained models in addressing geometric vision tasks.
A 28nm 0.22 μJ/token memory-compute-intensity-aware CNN-Transformer accelerator with hybrid-attention-based layer-fusion and cascaded pruning for semanticsegmentation
Dec 19, 2025Abstract:This work presents a 28nm 13.93mm2 CNN-Transformer accelerator for semantic segmentation, achieving 3.86-to-10.91x energy reduction over previous designs. It features a hybrid attention unit, layer-fusion scheduler, and cascaded feature-map pruner, with peak energy efficiency of 52.90TOPS/W (INT8).
* 3 pages,7 pages, 2025 IEEE International Solid-State Circuits Conference (ISSCC)
SR-LIO++: Efficient LiDAR-Inertial Odometry and Quantized Mapping with Sweep Reconstruction
Mar 29, 2025Abstract:Addressing the inherent low acquisition frequency limitation of 3D LiDAR to achieve high-frequency output has become a critical research focus in the LiDAR-Inertial Odometry (LIO) domain. To ensure real-time performance, frequency-enhanced LIO systems must process each sweep within significantly reduced timeframe, which presents substantial challenges for deployment on low-computational-power platforms. To address these limitations, we introduce SR-LIO++, an innovative LIO system capable of achieving doubled output frequency relative to input frequency on resource-constrained hardware platforms, including the Raspberry Pi 4B. Our system employs a sweep reconstruction methodology to enhance LiDAR sweep frequency, generating high-frequency reconstructed sweeps. Building upon this foundation, we propose a caching mechanism for intermediate results (i.e., surface parameters) of the most recent segments, effectively minimizing redundant processing of common segments in adjacent reconstructed sweeps. This method decouples processing time from the traditionally linear dependence on reconstructed sweep frequency. Furthermore, we present a quantized map point management based on index table mapping, significantly reducing memory usage by converting global 3D point storage from 64-bit double precision to 8-bit char representation. This method also converts the computationally intensive Euclidean distance calculations in nearest neighbor searches from 64-bit double precision to 16-bit short and 32-bit integer formats, significantly reducing both memory and computational cost. Extensive experimental evaluations across three distinct computing platforms and four public datasets demonstrate that SR-LIO++ maintains state-of-the-art accuracy while substantially enhancing efficiency. Notably, our system successfully achieves 20Hz state output on Raspberry Pi 4B hardware.
Memory Efficient Transformer Adapter for Dense Predictions
Feb 04, 2025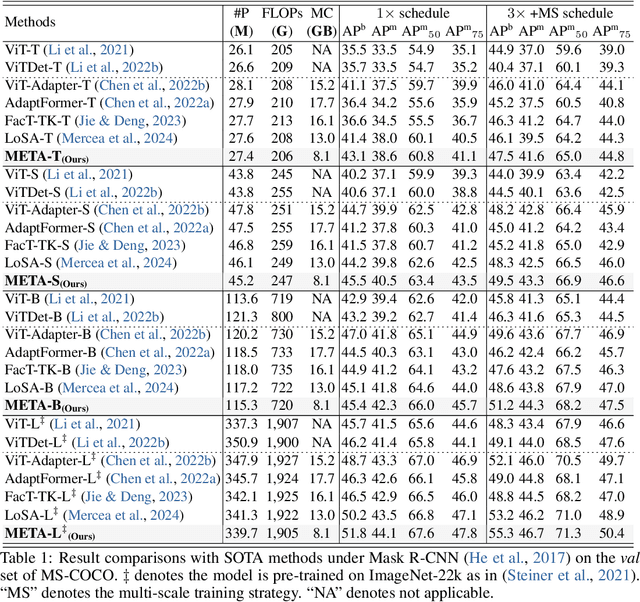
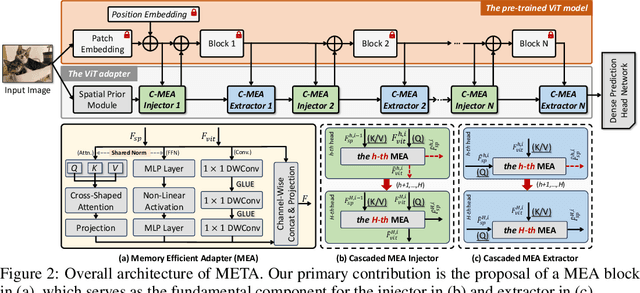
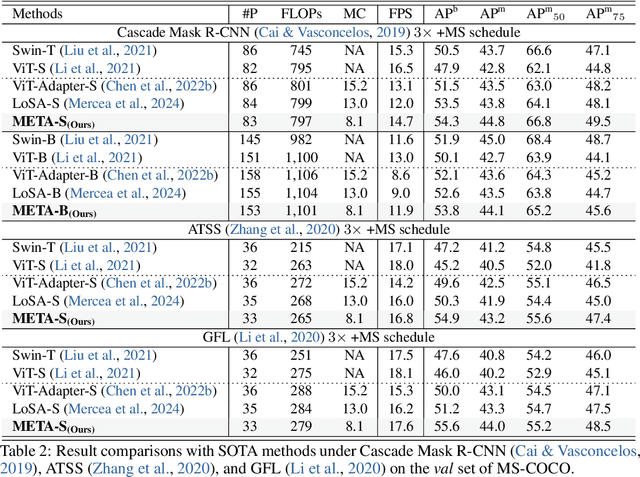
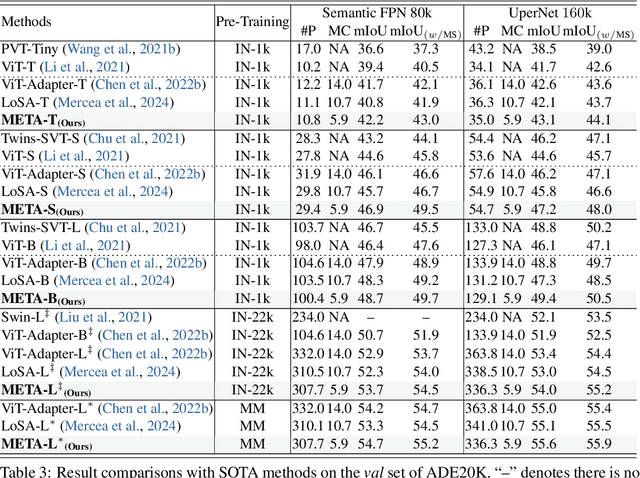
Abstract:While current Vision Transformer (ViT) adapter methods have shown promising accuracy, their inference speed is implicitly hindered by inefficient memory access operations, e.g., standard normalization and frequent reshaping. In this work, we propose META, a simple and fast ViT adapter that can improve the model's memory efficiency and decrease memory time consumption by reducing the inefficient memory access operations. Our method features a memory-efficient adapter block that enables the common sharing of layer normalization between the self-attention and feed-forward network layers, thereby reducing the model's reliance on normalization operations. Within the proposed block, the cross-shaped self-attention is employed to reduce the model's frequent reshaping operations. Moreover, we augment the adapter block with a lightweight convolutional branch that can enhance local inductive biases, particularly beneficial for the dense prediction tasks, e.g., object detection, instance segmentation, and semantic segmentation. The adapter block is finally formulated in a cascaded manner to compute diverse head features, thereby enriching the variety of feature representations. Empirically, extensive evaluations on multiple representative datasets validate that META substantially enhances the predicted quality, while achieving a new state-of-the-art accuracy-efficiency trade-off. Theoretically, we demonstrate that META exhibits superior generalization capability and stronger adaptability.
Generalized Task-Driven Medical Image Quality Enhancement with Gradient Promotion
Jan 02, 2025



Abstract:Thanks to the recent achievements in task-driven image quality enhancement (IQE) models like ESTR, the image enhancement model and the visual recognition model can mutually enhance each other's quantitation while producing high-quality processed images that are perceivable by our human vision systems. However, existing task-driven IQE models tend to overlook an underlying fact -- different levels of vision tasks have varying and sometimes conflicting requirements of image features. To address this problem, this paper proposes a generalized gradient promotion (GradProm) training strategy for task-driven IQE of medical images. Specifically, we partition a task-driven IQE system into two sub-models, i.e., a mainstream model for image enhancement and an auxiliary model for visual recognition. During training, GradProm updates only parameters of the image enhancement model using gradients of the visual recognition model and the image enhancement model, but only when gradients of these two sub-models are aligned in the same direction, which is measured by their cosine similarity. In case gradients of these two sub-models are not in the same direction, GradProm only uses the gradient of the image enhancement model to update its parameters. Theoretically, we have proved that the optimization direction of the image enhancement model will not be biased by the auxiliary visual recognition model under the implementation of GradProm. Empirically, extensive experimental results on four public yet challenging medical image datasets demonstrated the superior performance of GradProm over existing state-of-the-art methods.
SnapGen: Taming High-Resolution Text-to-Image Models for Mobile Devices with Efficient Architectures and Training
Dec 12, 2024Abstract:Existing text-to-image (T2I) diffusion models face several limitations, including large model sizes, slow runtime, and low-quality generation on mobile devices. This paper aims to address all of these challenges by developing an extremely small and fast T2I model that generates high-resolution and high-quality images on mobile platforms. We propose several techniques to achieve this goal. First, we systematically examine the design choices of the network architecture to reduce model parameters and latency, while ensuring high-quality generation. Second, to further improve generation quality, we employ cross-architecture knowledge distillation from a much larger model, using a multi-level approach to guide the training of our model from scratch. Third, we enable a few-step generation by integrating adversarial guidance with knowledge distillation. For the first time, our model SnapGen, demonstrates the generation of 1024x1024 px images on a mobile device around 1.4 seconds. On ImageNet-1K, our model, with only 372M parameters, achieves an FID of 2.06 for 256x256 px generation. On T2I benchmarks (i.e., GenEval and DPG-Bench), our model with merely 379M parameters, surpasses large-scale models with billions of parameters at a significantly smaller size (e.g., 7x smaller than SDXL, 14x smaller than IF-XL).
EoRA: Training-free Compensation for Compressed LLM with Eigenspace Low-Rank Approximation
Oct 28, 2024Abstract:In this work, we re-formulate the model compression problem into the customized compensation problem: Given a compressed model, we aim to introduce residual low-rank paths to compensate for compression errors under customized requirements from users (e.g., tasks, compression ratios), resulting in greater flexibility in adjusting overall capacity without being constrained by specific compression formats. However, naively applying SVD to derive residual paths causes suboptimal utilization of the low-rank representation capacity. Instead, we propose Training-free Eigenspace Low-Rank Approximation (EoRA), a method that directly minimizes compression-induced errors without requiring gradient-based training, achieving fast optimization in minutes using a small amount of calibration data. EoRA projects compression errors into the eigenspace of input activations, leveraging eigenvalues to effectively prioritize the reconstruction of high-importance error components. Moreover, EoRA can be seamlessly integrated with fine-tuning and quantization to further improve effectiveness and efficiency. EoRA consistently outperforms previous methods in compensating errors for compressed LLaMA2/3 models on various tasks, such as language generation, commonsense reasoning, and math reasoning tasks (e.g., 31.31%/12.88% and 9.69% improvements on ARC-Easy/ARC-Challenge and MathQA when compensating LLaMA3-8B that is quantized to 4-bit and pruned to 2:4 sparsity). EoRA offers a scalable, training-free solution to compensate for compression errors, making it a powerful tool to deploy LLMs in various capacity and efficiency requirements.
Revisiting Deep Ensemble Uncertainty for Enhanced Medical Anomaly Detection
Sep 26, 2024Abstract:Medical anomaly detection (AD) is crucial in pathological identification and localization. Current methods typically rely on uncertainty estimation in deep ensembles to detect anomalies, assuming that ensemble learners should agree on normal samples while exhibiting disagreement on unseen anomalies in the output space. However, these methods may suffer from inadequate disagreement on anomalies or diminished agreement on normal samples. To tackle these issues, we propose D2UE, a Diversified Dual-space Uncertainty Estimation framework for medical anomaly detection. To effectively balance agreement and disagreement for anomaly detection, we propose Redundancy-Aware Repulsion (RAR), which uses a similarity kernel that remains invariant to both isotropic scaling and orthogonal transformations, explicitly promoting diversity in learners' feature space. Moreover, to accentuate anomalous regions, we develop Dual-Space Uncertainty (DSU), which utilizes the ensemble's uncertainty in input and output spaces. In input space, we first calculate gradients of reconstruction error with respect to input images. The gradients are then integrated with reconstruction outputs to estimate uncertainty for inputs, enabling effective anomaly discrimination even when output space disagreement is minimal. We conduct a comprehensive evaluation of five medical benchmarks with different backbones. Experimental results demonstrate the superiority of our method to state-of-the-art methods and the effectiveness of each component in our framework. Our code is available at https://github.com/Rubiscol/D2UE.
Labeled-to-Unlabeled Distribution Alignment for Partially-Supervised Multi-Organ Medical Image Segmentation
Sep 05, 2024



Abstract:Partially-supervised multi-organ medical image segmentation aims to develop a unified semantic segmentation model by utilizing multiple partially-labeled datasets, with each dataset providing labels for a single class of organs. However, the limited availability of labeled foreground organs and the absence of supervision to distinguish unlabeled foreground organs from the background pose a significant challenge, which leads to a distribution mismatch between labeled and unlabeled pixels. Although existing pseudo-labeling methods can be employed to learn from both labeled and unlabeled pixels, they are prone to performance degradation in this task, as they rely on the assumption that labeled and unlabeled pixels have the same distribution. In this paper, to address the problem of distribution mismatch, we propose a labeled-to-unlabeled distribution alignment (LTUDA) framework that aligns feature distributions and enhances discriminative capability. Specifically, we introduce a cross-set data augmentation strategy, which performs region-level mixing between labeled and unlabeled organs to reduce distribution discrepancy and enrich the training set. Besides, we propose a prototype-based distribution alignment method that implicitly reduces intra-class variation and increases the separation between the unlabeled foreground and background. This can be achieved by encouraging consistency between the outputs of two prototype classifiers and a linear classifier. Extensive experimental results on the AbdomenCT-1K dataset and a union of four benchmark datasets (including LiTS, MSD-Spleen, KiTS, and NIH82) demonstrate that our method outperforms the state-of-the-art partially-supervised methods by a considerable margin, and even surpasses the fully-supervised methods. The source code is publicly available at https://github.com/xjiangmed/LTUDA.
Aligning Medical Images with General Knowledge from Large Language Models
Aug 31, 2024



Abstract:Pre-trained large vision-language models (VLMs) like CLIP have revolutionized visual representation learning using natural language as supervisions, and demonstrated promising generalization ability. In this work, we propose ViP, a novel visual symptom-guided prompt learning framework for medical image analysis, which facilitates general knowledge transfer from CLIP. ViP consists of two key components: a visual symptom generator (VSG) and a dual-prompt network. Specifically, VSG aims to extract explicable visual symptoms from pre-trained large language models, while the dual-prompt network utilizes these visual symptoms to guide the training on two learnable prompt modules, i.e., context prompt and merge prompt, which effectively adapts our framework to medical image analysis via large VLMs. Extensive experimental results demonstrate that ViP can outperform state-of-the-art methods on two challenging datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge