Yingxue Xu
A Deployment-Friendly Foundational Framework for Efficient Computational Pathology
Feb 15, 2026Abstract:Pathology foundation models (PFMs) have enabled robust generalization in computational pathology through large-scale datasets and expansive architectures, but their substantial computational cost, particularly for gigapixel whole slide images, limits clinical accessibility and scalability. Here, we present LitePath, a deployment-friendly foundational framework designed to mitigate model over-parameterization and patch level redundancy. LitePath integrates LiteFM, a compact model distilled from three large PFMs (Virchow2, H-Optimus-1 and UNI2) using 190 million patches, and the Adaptive Patch Selector (APS), a lightweight component for task-specific patch selection. The framework reduces model parameters by 28x and lowers FLOPs by 403.5x relative to Virchow2, enabling deployment on low-power edge hardware such as the NVIDIA Jetson Orin Nano Super. On this device, LitePath processes 208 slides per hour, 104.5x faster than Virchow2, and consumes 0.36 kWh per 3,000 slides, 171x lower than Virchow2 on an RTX3090 GPU. We validated accuracy using 37 cohorts across four organs and 26 tasks (26 internal, 9 external, and 2 prospective), comprising 15,672 slides from 9,808 patients disjoint from the pretraining data. LitePath ranks second among 19 evaluated models and outperforms larger models including H-Optimus-1, mSTAR, UNI2 and GPFM, while retaining 99.71% of the AUC of Virchow2 on average. To quantify the balance between accuracy and efficiency, we propose the Deployability Score (D-Score), defined as the weighted geometric mean of normalized AUC and normalized FLOP, where LitePath achieves the highest value, surpassing Virchow2 by 10.64%. These results demonstrate that LitePath enables rapid, cost-effective and energy-efficient pathology image analysis on accessible hardware while maintaining accuracy comparable to state-of-the-art PFMs and reducing the carbon footprint of AI deployment.
MambaMIL+: Modeling Long-Term Contextual Patterns for Gigapixel Whole Slide Image
Dec 19, 2025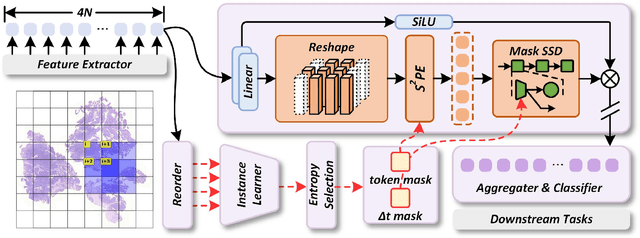
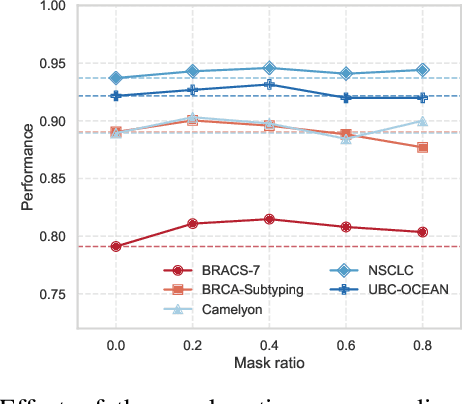
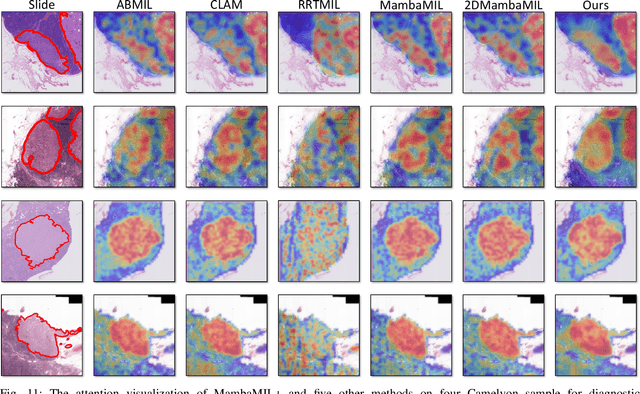
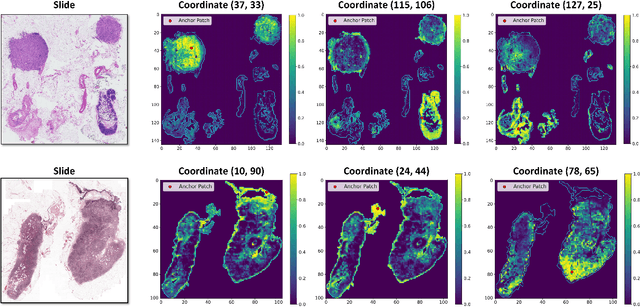
Abstract:Whole-slide images (WSIs) are an important data modality in computational pathology, yet their gigapixel resolution and lack of fine-grained annotations challenge conventional deep learning models. Multiple instance learning (MIL) offers a solution by treating each WSI as a bag of patch-level instances, but effectively modeling ultra-long sequences with rich spatial context remains difficult. Recently, Mamba has emerged as a promising alternative for long sequence learning, scaling linearly to thousands of tokens. However, despite its efficiency, it still suffers from limited spatial context modeling and memory decay, constraining its effectiveness to WSI analysis. To address these limitations, we propose MambaMIL+, a new MIL framework that explicitly integrates spatial context while maintaining long-range dependency modeling without memory forgetting. Specifically, MambaMIL+ introduces 1) overlapping scanning, which restructures the patch sequence to embed spatial continuity and instance correlations; 2) a selective stripe position encoder (S2PE) that encodes positional information while mitigating the biases of fixed scanning orders; and 3) a contextual token selection (CTS) mechanism, which leverages supervisory knowledge to dynamically enlarge the contextual memory for stable long-range modeling. Extensive experiments on 20 benchmarks across diagnostic classification, molecular prediction, and survival analysis demonstrate that MambaMIL+ consistently achieves state-of-the-art performance under three feature extractors (ResNet-50, PLIP, and CONCH), highlighting its effectiveness and robustness for large-scale computational pathology
LLM-driven Knowledge Enhancement for Multimodal Cancer Survival Prediction
Dec 16, 2025Abstract:Current multimodal survival prediction methods typically rely on pathology images (WSIs) and genomic data, both of which are high-dimensional and redundant, making it difficult to extract discriminative features from them and align different modalities. Moreover, using a simple survival follow-up label is insufficient to supervise such a complex task. To address these challenges, we propose KEMM, an LLM-driven Knowledge-Enhanced Multimodal Model for cancer survival prediction, which integrates expert reports and prognostic background knowledge. 1) Expert reports, provided by pathologists on a case-by-case basis and refined by large language model (LLM), offer succinct and clinically focused diagnostic statements. This information may typically suggest different survival outcomes. 2) Prognostic background knowledge (PBK), generated concisely by LLM, provides valuable prognostic background knowledge on different cancer types, which also enhances survival prediction. To leverage these knowledge, we introduce the knowledge-enhanced cross-modal (KECM) attention module. KECM can effectively guide the network to focus on discriminative and survival-relevant features from highly redundant modalities. Extensive experiments on five datasets demonstrate that KEMM achieves state-of-the-art performance. The code will be released upon acceptance.
GenAR: Next-Scale Autoregressive Generation for Spatial Gene Expression Prediction
Oct 05, 2025



Abstract:Spatial Transcriptomics (ST) offers spatially resolved gene expression but remains costly. Predicting expression directly from widely available Hematoxylin and Eosin (H&E) stained images presents a cost-effective alternative. However, most computational approaches (i) predict each gene independently, overlooking co-expression structure, and (ii) cast the task as continuous regression despite expression being discrete counts. This mismatch can yield biologically implausible outputs and complicate downstream analyses. We introduce GenAR, a multi-scale autoregressive framework that refines predictions from coarse to fine. GenAR clusters genes into hierarchical groups to expose cross-gene dependencies, models expression as codebook-free discrete token generation to directly predict raw counts, and conditions decoding on fused histological and spatial embeddings. From an information-theoretic perspective, the discrete formulation avoids log-induced biases and the coarse-to-fine factorization aligns with a principled conditional decomposition. Extensive experimental results on four Spatial Transcriptomics datasets across different tissue types demonstrate that GenAR achieves state-of-the-art performance, offering potential implications for precision medicine and cost-effective molecular profiling. Code is publicly available at https://github.com/oyjr/genar.
A Multimodal Foundation Model to Enhance Generalizability and Data Efficiency for Pan-cancer Prognosis Prediction
Sep 16, 2025Abstract:Multimodal data provides heterogeneous information for a holistic understanding of the tumor microenvironment. However, existing AI models often struggle to harness the rich information within multimodal data and extract poorly generalizable representations. Here we present MICE (Multimodal data Integration via Collaborative Experts), a multimodal foundation model that effectively integrates pathology images, clinical reports, and genomics data for precise pan-cancer prognosis prediction. Instead of conventional multi-expert modules, MICE employs multiple functionally diverse experts to comprehensively capture both cross-cancer and cancer-specific insights. Leveraging data from 11,799 patients across 30 cancer types, we enhanced MICE's generalizability by coupling contrastive and supervised learning. MICE outperformed both unimodal and state-of-the-art multi-expert-based multimodal models, demonstrating substantial improvements in C-index ranging from 3.8% to 11.2% on internal cohorts and 5.8% to 8.8% on independent cohorts, respectively. Moreover, it exhibited remarkable data efficiency across diverse clinical scenarios. With its enhanced generalizability and data efficiency, MICE establishes an effective and scalable foundation for pan-cancer prognosis prediction, holding strong potential to personalize tailored therapies and improve treatment outcomes.
A Versatile Pathology Co-pilot via Reasoning Enhanced Multimodal Large Language Model
Jul 23, 2025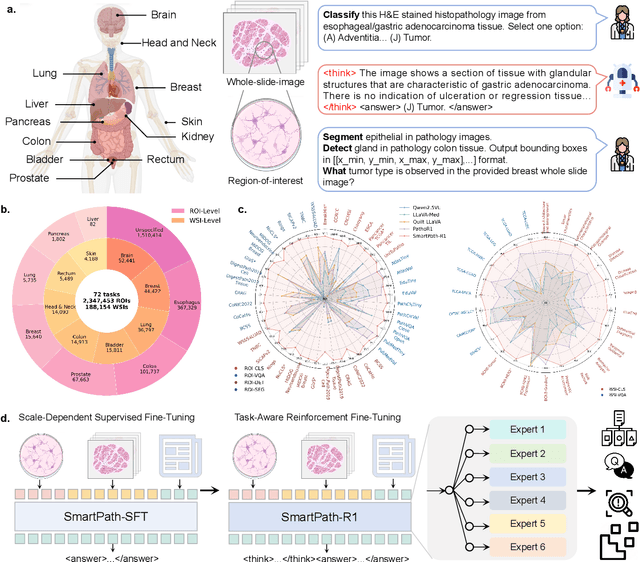
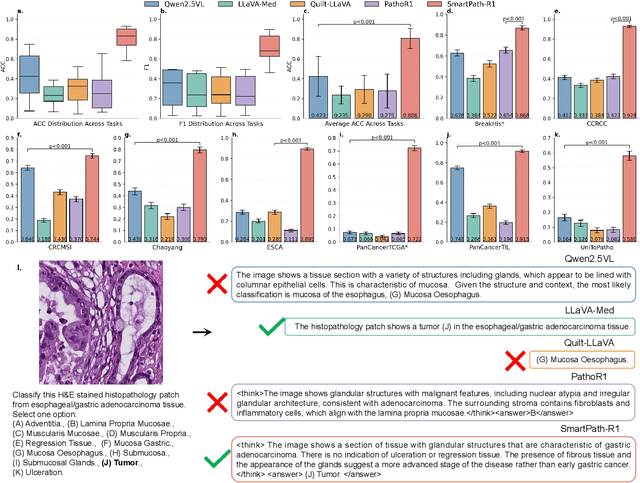
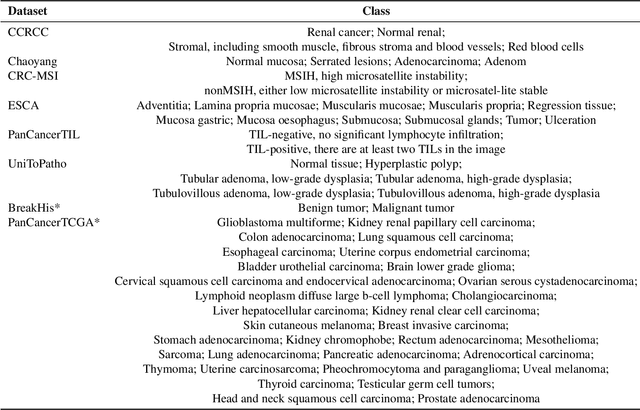
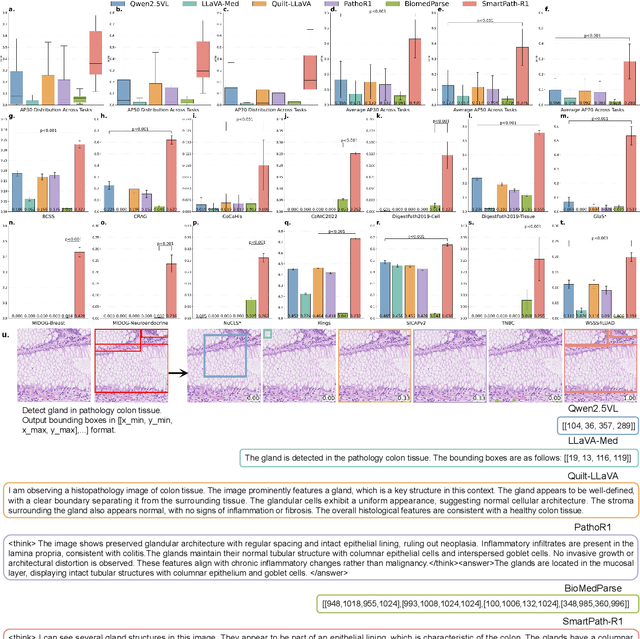
Abstract:Multimodal large language models (MLLMs) have emerged as powerful tools for computational pathology, offering unprecedented opportunities to integrate pathological images with language context for comprehensive diagnostic analysis. These models hold particular promise for automating complex tasks that traditionally require expert interpretation of pathologists. However, current MLLM approaches in pathology demonstrate significantly constrained reasoning capabilities, primarily due to their reliance on expensive chain-of-thought annotations. Additionally, existing methods remain limited to simplex application of visual question answering (VQA) at region-of-interest (ROI) level, failing to address the full spectrum of diagnostic needs such as ROI classification, detection, segmentation, whole-slide-image (WSI) classification and VQA in clinical practice. In this study, we present SmartPath-R1, a versatile MLLM capable of simultaneously addressing both ROI-level and WSI-level tasks while demonstrating robust pathological reasoning capability. Our framework combines scale-dependent supervised fine-tuning and task-aware reinforcement fine-tuning, which circumvents the requirement for chain-of-thought supervision by leveraging the intrinsic knowledge within MLLM. Furthermore, SmartPath-R1 integrates multiscale and multitask analysis through a mixture-of-experts mechanism, enabling dynamic processing for diverse tasks. We curate a large-scale dataset comprising 2.3M ROI samples and 188K WSI samples for training and evaluation. Extensive experiments across 72 tasks validate the effectiveness and superiority of the proposed approach. This work represents a significant step toward developing versatile, reasoning-enhanced AI systems for precision pathology.
Genome-Anchored Foundation Model Embeddings Improve Molecular Prediction from Histology Images
Jun 24, 2025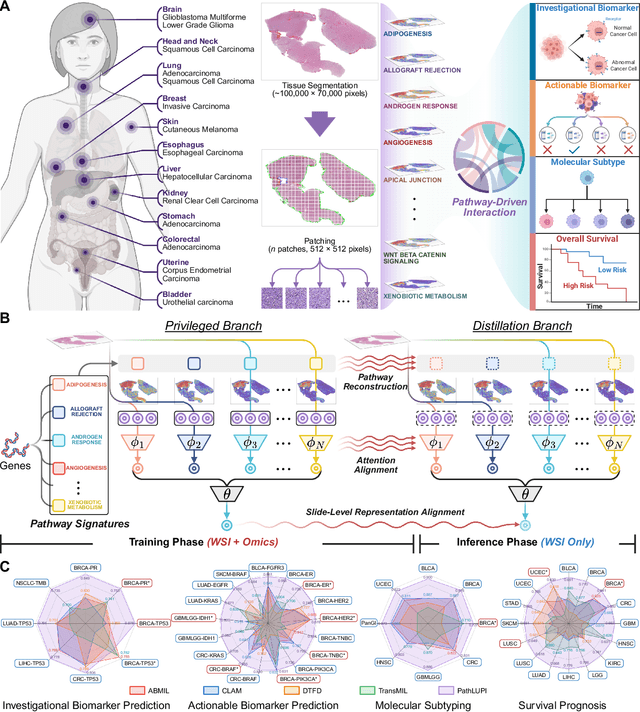
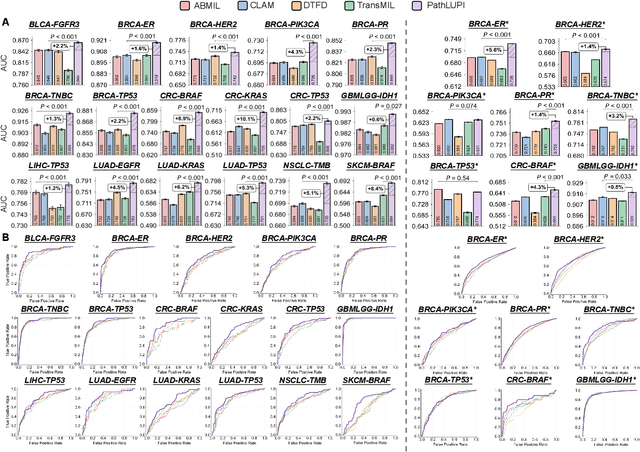
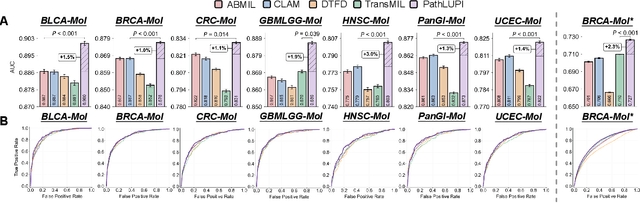
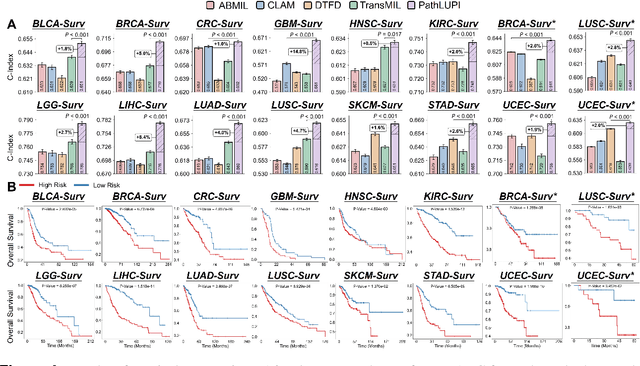
Abstract:Precision oncology requires accurate molecular insights, yet obtaining these directly from genomics is costly and time-consuming for broad clinical use. Predicting complex molecular features and patient prognosis directly from routine whole-slide images (WSI) remains a major challenge for current deep learning methods. Here we introduce PathLUPI, which uses transcriptomic privileged information during training to extract genome-anchored histological embeddings, enabling effective molecular prediction using only WSIs at inference. Through extensive evaluation across 49 molecular oncology tasks using 11,257 cases among 20 cohorts, PathLUPI demonstrated superior performance compared to conventional methods trained solely on WSIs. Crucially, it achieves AUC $\geq$ 0.80 in 14 of the biomarker prediction and molecular subtyping tasks and C-index $\geq$ 0.70 in survival cohorts of 5 major cancer types. Moreover, PathLUPI embeddings reveal distinct cellular morphological signatures associated with specific genotypes and related biological pathways within WSIs. By effectively encoding molecular context to refine WSI representations, PathLUPI overcomes a key limitation of existing models and offers a novel strategy to bridge molecular insights with routine pathology workflows for wider clinical application.
PathBench: A comprehensive comparison benchmark for pathology foundation models towards precision oncology
May 26, 2025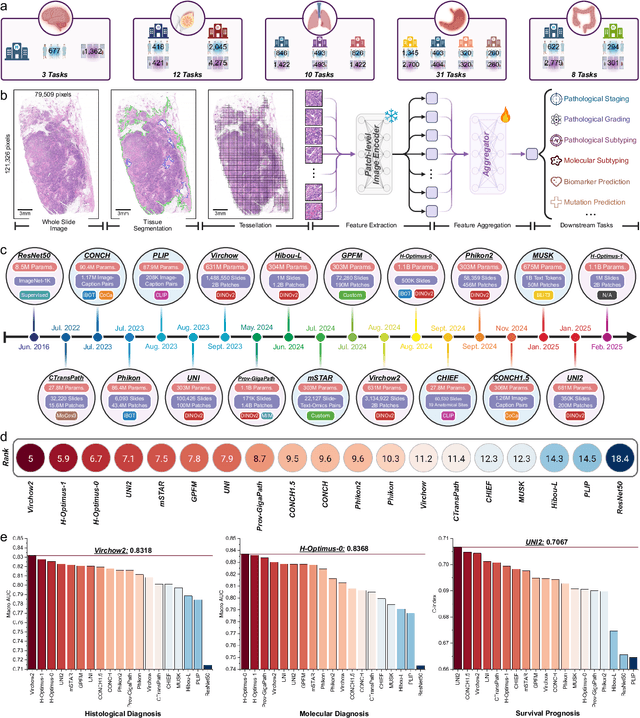



Abstract:The emergence of pathology foundation models has revolutionized computational histopathology, enabling highly accurate, generalized whole-slide image analysis for improved cancer diagnosis, and prognosis assessment. While these models show remarkable potential across cancer diagnostics and prognostics, their clinical translation faces critical challenges including variability in optimal model across cancer types, potential data leakage in evaluation, and lack of standardized benchmarks. Without rigorous, unbiased evaluation, even the most advanced PFMs risk remaining confined to research settings, delaying their life-saving applications. Existing benchmarking efforts remain limited by narrow cancer-type focus, potential pretraining data overlaps, or incomplete task coverage. We present PathBench, the first comprehensive benchmark addressing these gaps through: multi-center in-hourse datasets spanning common cancers with rigorous leakage prevention, evaluation across the full clinical spectrum from diagnosis to prognosis, and an automated leaderboard system for continuous model assessment. Our framework incorporates large-scale data, enabling objective comparison of PFMs while reflecting real-world clinical complexity. All evaluation data comes from private medical providers, with strict exclusion of any pretraining usage to avoid data leakage risks. We have collected 15,888 WSIs from 8,549 patients across 10 hospitals, encompassing over 64 diagnosis and prognosis tasks. Currently, our evaluation of 19 PFMs shows that Virchow2 and H-Optimus-1 are the most effective models overall. This work provides researchers with a robust platform for model development and offers clinicians actionable insights into PFM performance across diverse clinical scenarios, ultimately accelerating the translation of these transformative technologies into routine pathology practice.
Distilled Prompt Learning for Incomplete Multimodal Survival Prediction
Mar 03, 2025Abstract:The integration of multimodal data including pathology images and gene profiles is widely applied in precise survival prediction. Despite recent advances in multimodal survival models, collecting complete modalities for multimodal fusion still poses a significant challenge, hindering their application in clinical settings. Current approaches tackling incomplete modalities often fall short, as they typically compensate for only a limited part of the knowledge of missing modalities. To address this issue, we propose a Distilled Prompt Learning framework (DisPro) to utilize the strong robustness of Large Language Models (LLMs) to missing modalities, which employs two-stage prompting for compensation of comprehensive information for missing modalities. In the first stage, Unimodal Prompting (UniPro) distills the knowledge distribution of each modality, preparing for supplementing modality-specific knowledge of the missing modality in the subsequent stage. In the second stage, Multimodal Prompting (MultiPro) leverages available modalities as prompts for LLMs to infer the missing modality, which provides modality-common information. Simultaneously, the unimodal knowledge acquired in the first stage is injected into multimodal inference to compensate for the modality-specific knowledge of the missing modality. Extensive experiments covering various missing scenarios demonstrated the superiority of the proposed method. The code is available at https://github.com/Innse/DisPro.
Towards A Generalizable Pathology Foundation Model via Unified Knowledge Distillation
Jul 26, 2024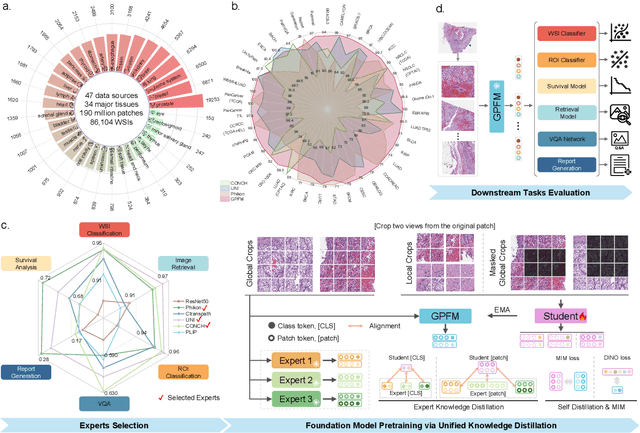

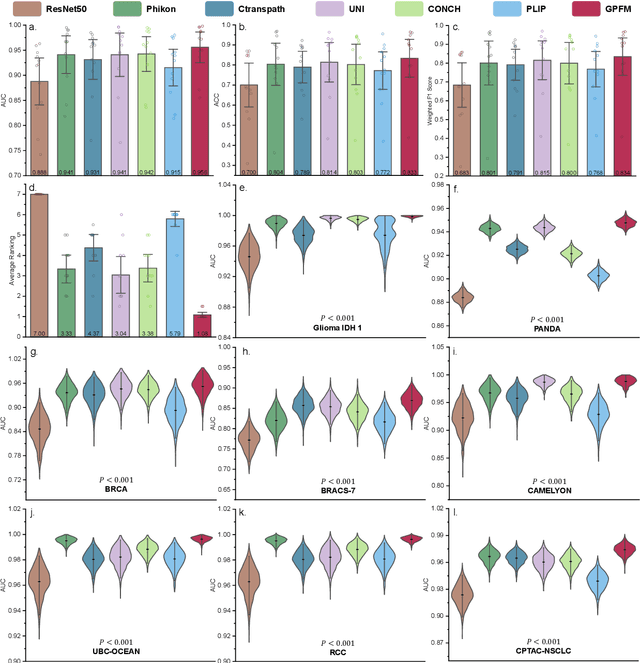
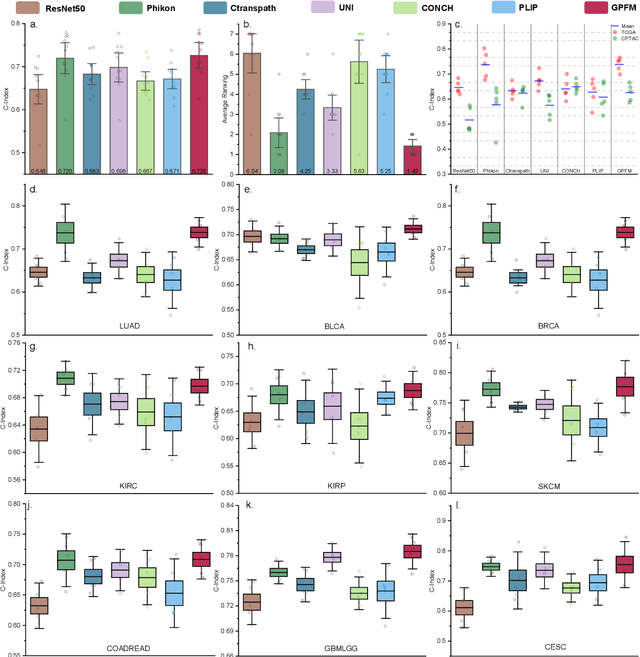
Abstract:Foundation models pretrained on large-scale datasets are revolutionizing the field of computational pathology (CPath). The generalization ability of foundation models is crucial for the success in various downstream clinical tasks. However, current foundation models have only been evaluated on a limited type and number of tasks, leaving their generalization ability and overall performance unclear. To address this gap, we established a most comprehensive benchmark to evaluate the performance of off-the-shelf foundation models across six distinct clinical task types, encompassing a total of 39 specific tasks. Our findings reveal that existing foundation models excel at certain task types but struggle to effectively handle the full breadth of clinical tasks. To improve the generalization of pathology foundation models, we propose a unified knowledge distillation framework consisting of both expert and self knowledge distillation, where the former allows the model to learn from the knowledge of multiple expert models, while the latter leverages self-distillation to enable image representation learning via local-global alignment. Based on this framework, a Generalizable Pathology Foundation Model (GPFM) is pretrained on a large-scale dataset consisting of 190 million images from around 86,000 public H\&E whole slides across 34 major tissue types. Evaluated on the established benchmark, GPFM achieves an impressive average rank of 1.36, with 29 tasks ranked 1st, while the the second-best model, UNI, attains an average rank of 2.96, with only 4 tasks ranked 1st. The superior generalization of GPFM demonstrates its exceptional modeling capabilities across a wide range of clinical tasks, positioning it as a new cornerstone for feature representation in CPath.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge