Zhixuan Chen
SAES-SVD: Self-Adaptive Suppression of Accumulated and Local Errors for SVD-based LLM Compression
Feb 03, 2026Abstract:The rapid growth in the parameter scale of large language models (LLMs) has created a high demand for efficient compression techniques. As a hardware-agnostic and highly compatible technique, low-rank compression has been widely adopted. However, existing methods typically compress each layer independently by minimizing per-layer reconstruction error, overlooking a critical limitation: the reconstruction error propagates and accumulates through the network, which leads to amplified global deviations from the full-precision baseline. To address this, we propose Self-Adaptive Error Suppression SVD (SAES-SVD), a LLMs compression framework that jointly optimizes intra-layer reconstruction and inter-layer error compensation. SAES-SVD is composed of two novel components: (1) Cumulative Error-Aware Layer Compression (CEALC), which formulates the compression objective as a combination of local reconstruction and weighted cumulative error compensation. Based on it, we derive a closed-form low-rank solution relied on second-order activation statistics, which explicitly aligns each layer's output with its full-precision counterpart to compensate for accumulated errors. (2) Adaptive Collaborative Error Suppression (ACES), which automatically adjusts the weighting coefficient to enhance the low-rank structure of the compression objective in CEALC. Specifically, the coefficient is optimized to maximize the ratio between the Frobenius norm of the compressed layer's output and that of the compression objective under a fixed rank, thus ensuring that the rank budget is utilized effectively. Extensive experiments across multiple LLM architectures and tasks show that, without fine-tuning or mixed-rank strategies, SAES-SVD consistently improves post-compression performance.
OTARo: Once Tuning for All Precisions toward Robust On-Device LLMs
Nov 17, 2025Abstract:Large Language Models (LLMs) fine-tuning techniques not only improve the adaptability to diverse downstream tasks, but also mitigate adverse effects of model quantization. Despite this, conventional quantization suffers from its structural limitation that hinders flexibility during the fine-tuning and deployment stages. Practical on-device tasks demand different quantization precisions (i.e. different bit-widths), e.g., understanding tasks tend to exhibit higher tolerance to reduced precision compared to generation tasks. Conventional quantization, typically relying on scaling factors that are incompatible across bit-widths, fails to support the on-device switching of precisions when confronted with complex real-world scenarios. To overcome the dilemma, we propose OTARo, a novel method that enables on-device LLMs to flexibly switch quantization precisions while maintaining performance robustness through once fine-tuning. OTARo introduces Shared Exponent Floating Point (SEFP), a distinct quantization mechanism, to produce different bit-widths through simple mantissa truncations of a single model. Moreover, to achieve bit-width robustness in downstream applications, OTARo performs a learning process toward losses induced by different bit-widths. The method involves two critical strategies: (1) Exploitation-Exploration Bit-Width Path Search (BPS), which iteratively updates the search path via a designed scoring mechanism; (2) Low-Precision Asynchronous Accumulation (LAA), which performs asynchronous gradient accumulations and delayed updates under low bit-widths. Experiments on popular LLMs, e.g., LLaMA3.2-1B, LLaMA3-8B, demonstrate that OTARo achieves consistently strong and robust performance for all precisions.
DiscoX: Benchmarking Discourse-Level Translation task in Expert Domains
Nov 14, 2025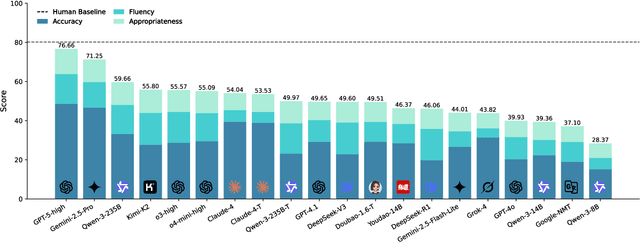
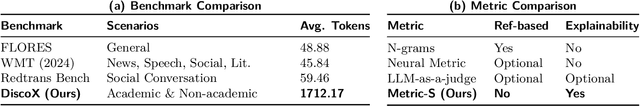
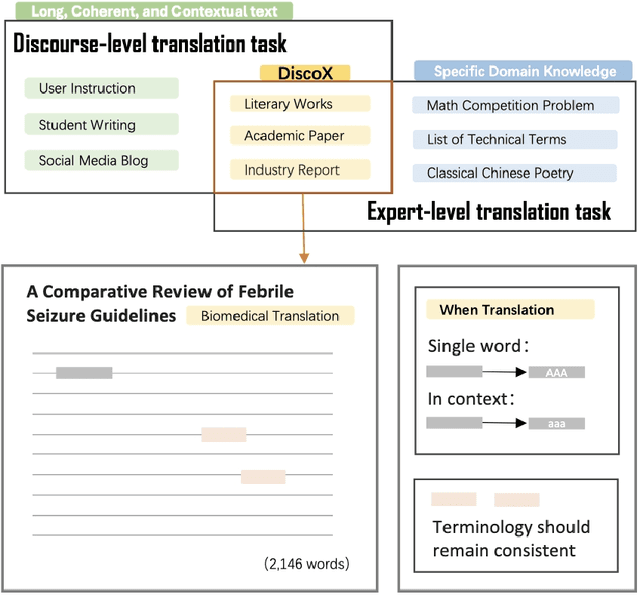
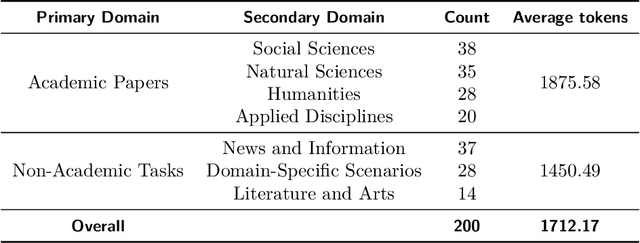
Abstract:The evaluation of discourse-level translation in expert domains remains inadequate, despite its centrality to knowledge dissemination and cross-lingual scholarly communication. While these translations demand discourse-level coherence and strict terminological precision, current evaluation methods predominantly focus on segment-level accuracy and fluency. To address this limitation, we introduce DiscoX, a new benchmark for discourse-level and expert-level Chinese-English translation. It comprises 200 professionally-curated texts from 7 domains, with an average length exceeding 1700 tokens. To evaluate performance on DiscoX, we also develop Metric-S, a reference-free system that provides fine-grained automatic assessments across accuracy, fluency, and appropriateness. Metric-S demonstrates strong consistency with human judgments, significantly outperforming existing metrics. Our experiments reveal a remarkable performance gap: even the most advanced LLMs still trail human experts on these tasks. This finding validates the difficulty of DiscoX and underscores the challenges that remain in achieving professional-grade machine translation. The proposed benchmark and evaluation system provide a robust framework for more rigorous evaluation, facilitating future advancements in LLM-based translation.
A Versatile Pathology Co-pilot via Reasoning Enhanced Multimodal Large Language Model
Jul 23, 2025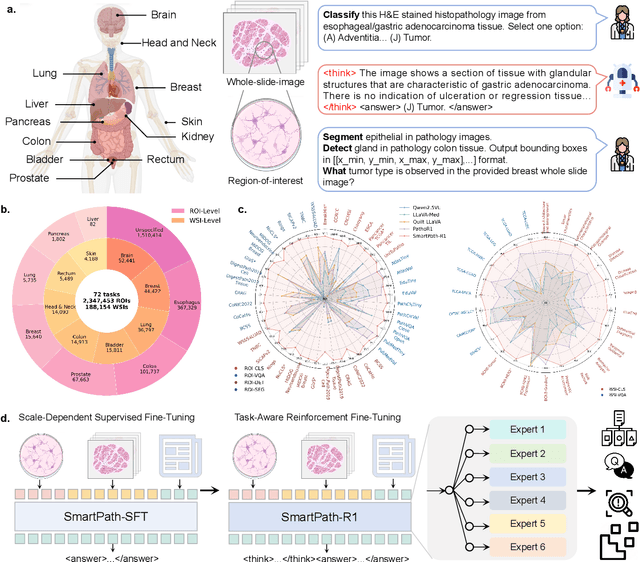
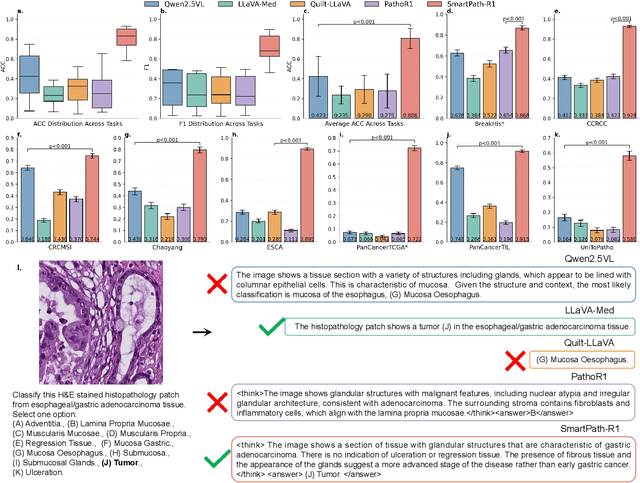
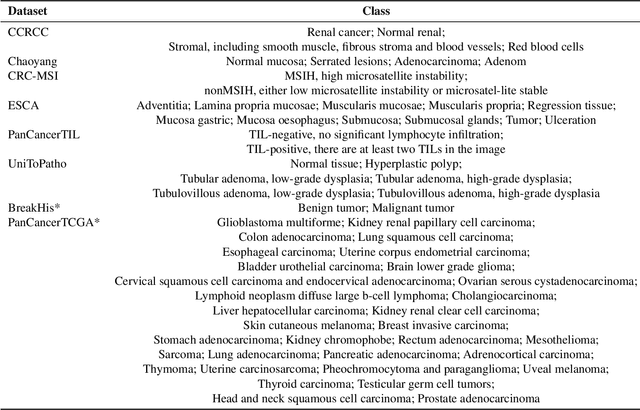
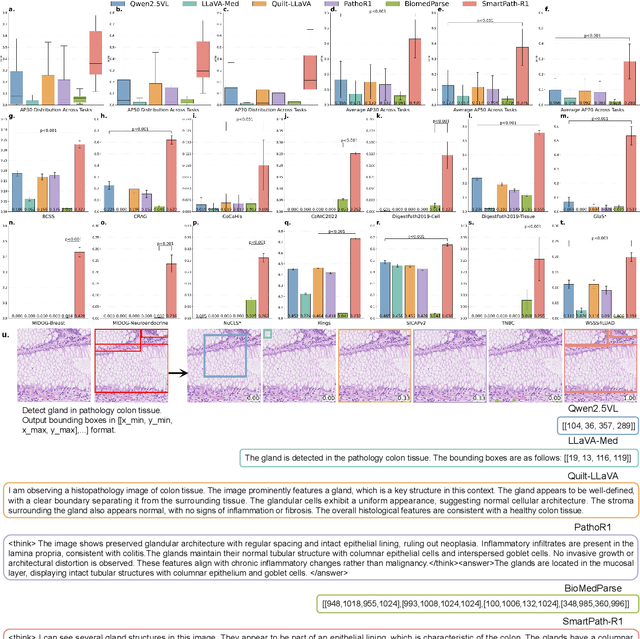
Abstract:Multimodal large language models (MLLMs) have emerged as powerful tools for computational pathology, offering unprecedented opportunities to integrate pathological images with language context for comprehensive diagnostic analysis. These models hold particular promise for automating complex tasks that traditionally require expert interpretation of pathologists. However, current MLLM approaches in pathology demonstrate significantly constrained reasoning capabilities, primarily due to their reliance on expensive chain-of-thought annotations. Additionally, existing methods remain limited to simplex application of visual question answering (VQA) at region-of-interest (ROI) level, failing to address the full spectrum of diagnostic needs such as ROI classification, detection, segmentation, whole-slide-image (WSI) classification and VQA in clinical practice. In this study, we present SmartPath-R1, a versatile MLLM capable of simultaneously addressing both ROI-level and WSI-level tasks while demonstrating robust pathological reasoning capability. Our framework combines scale-dependent supervised fine-tuning and task-aware reinforcement fine-tuning, which circumvents the requirement for chain-of-thought supervision by leveraging the intrinsic knowledge within MLLM. Furthermore, SmartPath-R1 integrates multiscale and multitask analysis through a mixture-of-experts mechanism, enabling dynamic processing for diverse tasks. We curate a large-scale dataset comprising 2.3M ROI samples and 188K WSI samples for training and evaluation. Extensive experiments across 72 tasks validate the effectiveness and superiority of the proposed approach. This work represents a significant step toward developing versatile, reasoning-enhanced AI systems for precision pathology.
Segment Anything in Pathology Images with Natural Language
Jun 26, 2025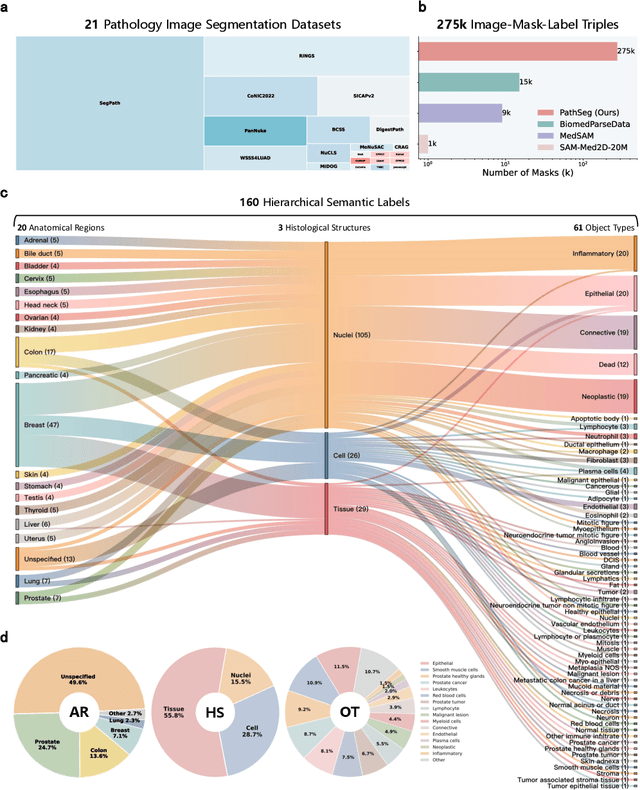
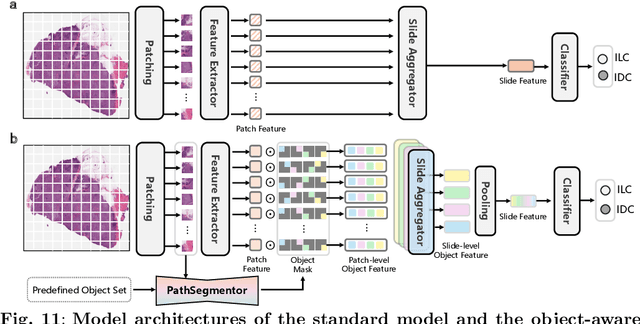
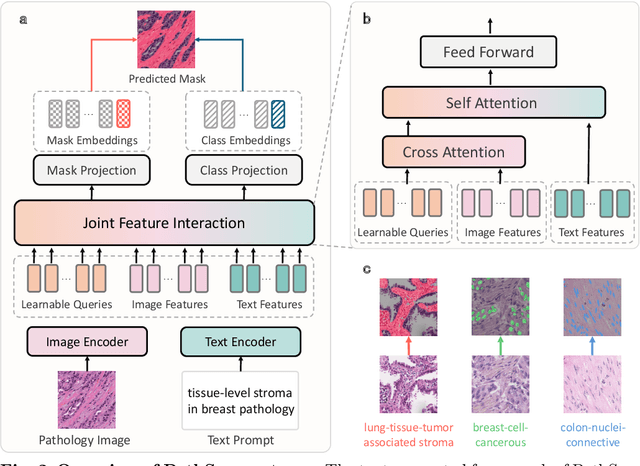
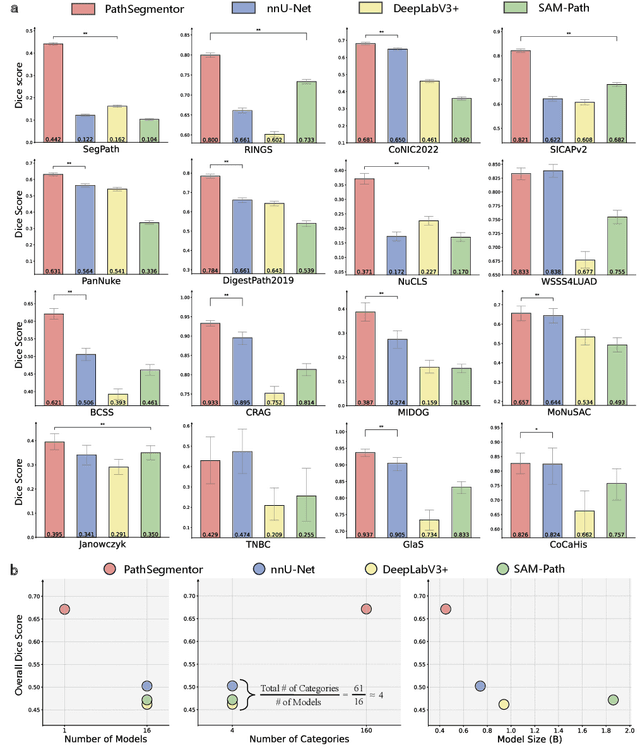
Abstract:Pathology image segmentation is crucial in computational pathology for analyzing histological features relevant to cancer diagnosis and prognosis. However, current methods face major challenges in clinical applications due to limited annotated data and restricted category definitions. To address these limitations, we propose PathSegmentor, the first text-prompted segmentation foundation model designed specifically for pathology images. We also introduce PathSeg , the largest and most comprehensive dataset for pathology segmentation, built from 17 public sources and containing 275k image-mask-label triples across 160 diverse categories. With PathSegmentor, users can perform semantic segmentation using natural language prompts, eliminating the need for laborious spatial inputs such as points or boxes. Extensive experiments demonstrate that PathSegmentor outperforms specialized models with higher accuracy and broader applicability, while maintaining a compact architecture. It significantly surpasses existing spatial- and text-prompted models by 0.145 and 0.429 in overall Dice scores, respectively, showing strong robustness in segmenting complex structures and generalizing to external datasets. Moreover, PathSegmentor's outputs enhance the interpretability of diagnostic models through feature importance estimation and imaging biomarker discovery, offering pathologists evidence-based support for clinical decision-making. This work advances the development of explainable AI in precision oncology.
RWKVQuant: Quantizing the RWKV Family with Proxy Guided Hybrid of Scalar and Vector Quantization
May 02, 2025



Abstract:RWKV is a modern RNN architecture with comparable performance to Transformer, but still faces challenges when deployed to resource-constrained devices. Post Training Quantization (PTQ), which is a an essential technique to reduce model size and inference latency, has been widely used in Transformer models. However, it suffers significant degradation of performance when applied to RWKV. This paper investigates and identifies two key constraints inherent in the properties of RWKV: (1) Non-linear operators hinder the parameter-fusion of both smooth- and rotation-based quantization, introducing extra computation overhead. (2) The larger amount of uniformly distributed weights poses challenges for cluster-based quantization, leading to reduced accuracy. To this end, we propose RWKVQuant, a PTQ framework tailored for RWKV models, consisting of two novel techniques: (1) a coarse-to-fine proxy capable of adaptively selecting different quantization approaches by assessing the uniformity and identifying outliers in the weights, and (2) a codebook optimization algorithm that enhances the performance of cluster-based quantization methods for element-wise multiplication in RWKV. Experiments show that RWKVQuant can quantize RWKV-6-14B into about 3-bit with less than 1% accuracy loss and 2.14x speed up.
MoEQuant: Enhancing Quantization for Mixture-of-Experts Large Language Models via Expert-Balanced Sampling and Affinity Guidance
May 02, 2025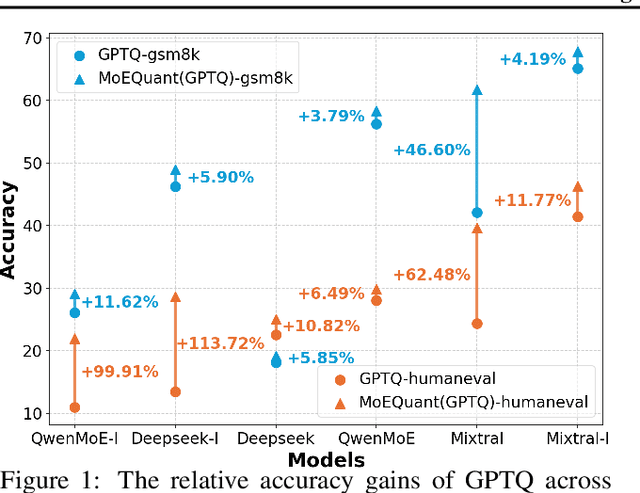
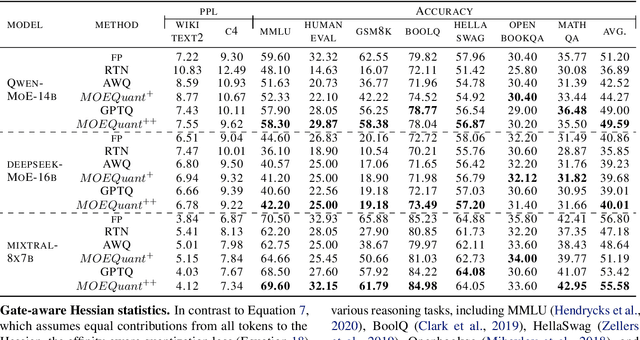
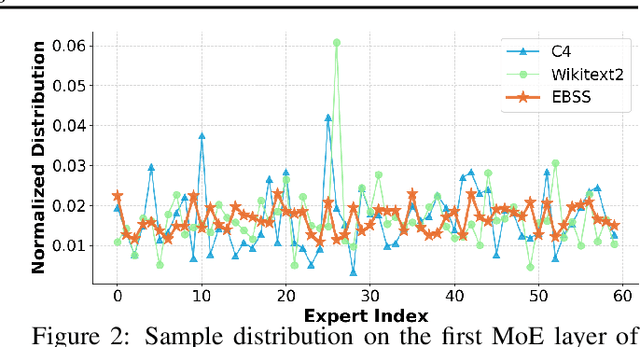
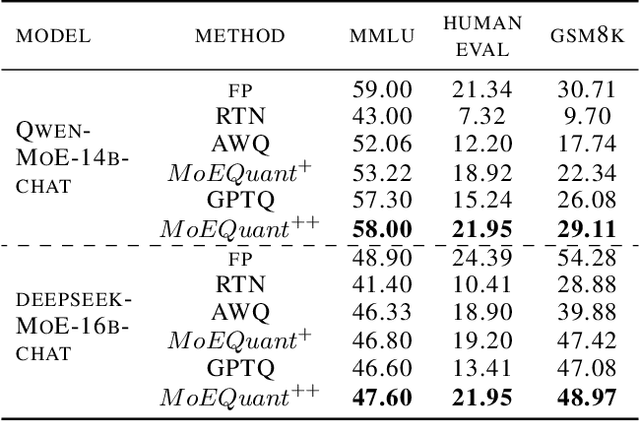
Abstract:Mixture-of-Experts (MoE) large language models (LLMs), which leverage dynamic routing and sparse activation to enhance efficiency and scalability, have achieved higher performance while reducing computational costs. However, these models face significant memory overheads, limiting their practical deployment and broader adoption. Post-training quantization (PTQ), a widely used method for compressing LLMs, encounters severe accuracy degradation and diminished generalization performance when applied to MoE models. This paper investigates the impact of MoE's sparse and dynamic characteristics on quantization and identifies two primary challenges: (1) Inter-expert imbalance, referring to the uneven distribution of samples across experts, which leads to insufficient and biased calibration for less frequently utilized experts; (2) Intra-expert imbalance, arising from MoE's unique aggregation mechanism, which leads to varying degrees of correlation between different samples and their assigned experts. To address these challenges, we propose MoEQuant, a novel quantization framework tailored for MoE LLMs. MoE-Quant includes two novel techniques: 1) Expert-Balanced Self-Sampling (EBSS) is an efficient sampling method that efficiently constructs a calibration set with balanced expert distributions by leveraging the cumulative probabilities of tokens and expert balance metrics as guiding factors. 2) Affinity-Guided Quantization (AGQ), which incorporates affinities between experts and samples into the quantization process, thereby accurately assessing the impact of individual samples on different experts within the MoE layer. Experiments demonstrate that MoEQuant achieves substantial performance gains (more than 10 points accuracy gain in the HumanEval for DeepSeekMoE-16B under 4-bit quantization) and boosts efficiency.
ConceptCLIP: Towards Trustworthy Medical AI via Concept-Enhanced Contrastive Langauge-Image Pre-training
Jan 26, 2025Abstract:Trustworthiness is essential for the precise and interpretable application of artificial intelligence (AI) in medical imaging. Traditionally, precision and interpretability have been addressed as separate tasks, namely medical image analysis and explainable AI, each developing its own models independently. In this study, for the first time, we investigate the development of a unified medical vision-language pre-training model that can achieve both accurate analysis and interpretable understanding of medical images across various modalities. To build the model, we construct MedConcept-23M, a large-scale dataset comprising 23 million medical image-text pairs extracted from 6.2 million scientific articles, enriched with concepts from the Unified Medical Language System (UMLS). Based on MedConcept-23M, we introduce ConceptCLIP, a medical AI model utilizing concept-enhanced contrastive language-image pre-training. The pre-training of ConceptCLIP involves two primary components: image-text alignment learning (IT-Align) and patch-concept alignment learning (PC-Align). This dual alignment strategy enhances the model's capability to associate specific image regions with relevant concepts, thereby improving both the precision of analysis and the interpretability of the AI system. We conducted extensive experiments on 5 diverse types of medical image analysis tasks, spanning 51 subtasks across 10 image modalities, with the broadest range of downstream tasks. The results demonstrate the effectiveness of the proposed vision-language pre-training model. Further explainability analysis across 6 modalities reveals that ConceptCLIP achieves superior performance, underscoring its robust ability to advance explainable AI in medical imaging. These findings highlight ConceptCLIP's capability in promoting trustworthy AI in the field of medicine.
Large Language Model with Region-guided Referring and Grounding for CT Report Generation
Nov 23, 2024



Abstract:Computed tomography (CT) report generation is crucial to assist radiologists in interpreting CT volumes, which can be time-consuming and labor-intensive. Existing methods primarily only consider the global features of the entire volume, making it struggle to focus on specific regions and potentially missing abnormalities. To address this issue, we propose Reg2RG, the first region-guided referring and grounding framework for CT report generation, which enhances diagnostic performance by focusing on anatomical regions within the volume. Specifically, we utilize masks from a universal segmentation module to capture local features for each referring region. A local feature decoupling (LFD) strategy is proposed to preserve the local high-resolution details with little computational overhead. Then the local features are integrated with global features to capture inter-regional relationships within a cohesive context. Moreover, we propose a novel region-report alignment (RRA) training strategy. It leverages the recognition of referring regions to guide the generation of region-specific reports, enhancing the model's referring and grounding capabilities while also improving the report's interpretability. A large language model (LLM) is further employed as the language decoder to generate reports from integrated visual features, facilitating region-level comprehension. Extensive experiments on two large-scale chest CT-report datasets demonstrate the superiority of our method, which outperforms several state-of-the-art methods in terms of both natural language generation and clinical efficacy metrics while preserving promising interpretability. The code will be made publicly available.
MedDr: Diagnosis-Guided Bootstrapping for Large-Scale Medical Vision-Language Learning
Apr 23, 2024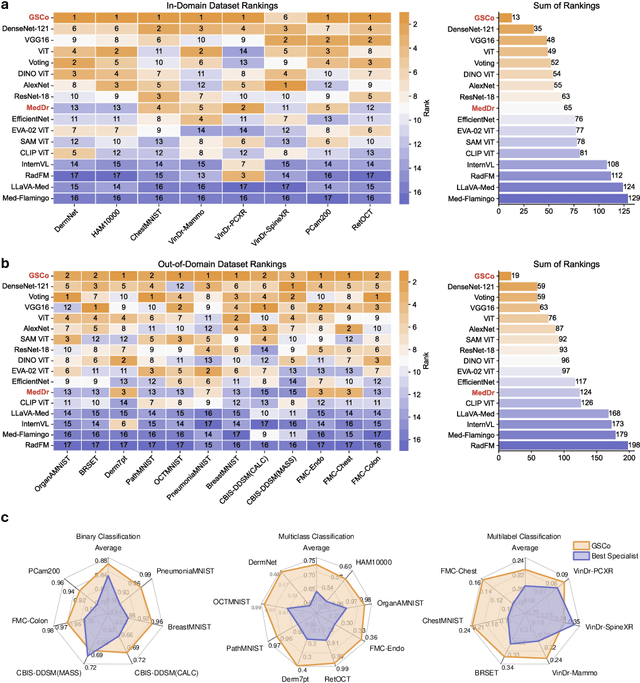

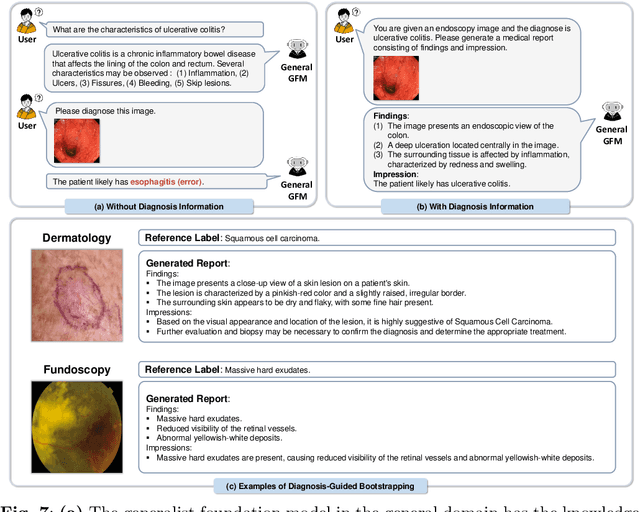
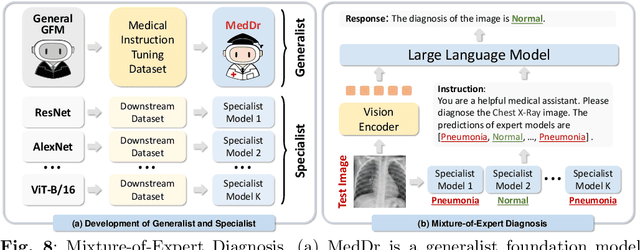
Abstract:The rapid advancement of large-scale vision-language models has showcased remarkable capabilities across various tasks. However, the lack of extensive and high-quality image-text data in medicine has greatly hindered the development of large-scale medical vision-language models. In this work, we present a diagnosis-guided bootstrapping strategy that exploits both image and label information to construct vision-language datasets. Based on the constructed dataset, we developed MedDr, a generalist foundation model for healthcare capable of handling diverse medical data modalities, including radiology, pathology, dermatology, retinography, and endoscopy. Moreover, during inference, we propose a simple but effective retrieval-augmented medical diagnosis strategy, which enhances the model's generalization ability. Extensive experiments on visual question answering, medical report generation, and medical image diagnosis demonstrate the superiority of our method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge