Jiaxin Zhuang
Glance and Focus Reinforcement for Pan-cancer Screening
Jan 27, 2026Abstract:Pan-cancer screening in large-scale CT scans remains challenging for existing AI methods, primarily due to the difficulty of localizing diverse types of tiny lesions in large CT volumes. The extreme foreground-background imbalance significantly hinders models from focusing on diseased regions, while redundant focus on healthy regions not only decreases the efficiency but also increases false positives. Inspired by radiologists' glance and focus diagnostic strategy, we introduce GF-Screen, a Glance and Focus reinforcement learning framework for pan-cancer screening. GF-Screen employs a Glance model to localize the diseased regions and a Focus model to precisely segment the lesions, where segmentation results of the Focus model are leveraged to reward the Glance model via Reinforcement Learning (RL). Specifically, the Glance model crops a group of sub-volumes from the entire CT volume and learns to select the sub-volumes with lesions for the Focus model to segment. Given that the selecting operation is non-differentiable for segmentation training, we propose to employ the segmentation results to reward the Glance model. To optimize the Glance model, we introduce a novel group relative learning paradigm, which employs group relative comparison to prioritize high-advantage predictions and discard low-advantage predictions within sub-volume groups, not only improving efficiency but also reducing false positives. In this way, for the first time, we effectively extend cutting-edge RL techniques to tackle the specific challenges in pan-cancer screening. Extensive experiments on 16 internal and 7 external datasets across 9 lesion types demonstrated the effectiveness of GF-Screen. Notably, GF-Screen leads the public validation leaderboard of MICCAI FLARE25 pan-cancer challenge, surpassing the FLARE24 champion solution by a large margin (+25.6% DSC and +28.2% NSD).
UniBiomed: A Universal Foundation Model for Grounded Biomedical Image Interpretation
Apr 30, 2025Abstract:Multi-modal interpretation of biomedical images opens up novel opportunities in biomedical image analysis. Conventional AI approaches typically rely on disjointed training, i.e., Large Language Models (LLMs) for clinical text generation and segmentation models for target extraction, which results in inflexible real-world deployment and a failure to leverage holistic biomedical information. To this end, we introduce UniBiomed, the first universal foundation model for grounded biomedical image interpretation. UniBiomed is based on a novel integration of Multi-modal Large Language Model (MLLM) and Segment Anything Model (SAM), which effectively unifies the generation of clinical texts and the segmentation of corresponding biomedical objects for grounded interpretation. In this way, UniBiomed is capable of tackling a wide range of biomedical tasks across ten diverse biomedical imaging modalities. To develop UniBiomed, we curate a large-scale dataset comprising over 27 million triplets of images, annotations, and text descriptions across ten imaging modalities. Extensive validation on 84 internal and external datasets demonstrated that UniBiomed achieves state-of-the-art performance in segmentation, disease recognition, region-aware diagnosis, visual question answering, and report generation. Moreover, unlike previous models that rely on clinical experts to pre-diagnose images and manually craft precise textual or visual prompts, UniBiomed can provide automated and end-to-end grounded interpretation for biomedical image analysis. This represents a novel paradigm shift in clinical workflows, which will significantly improve diagnostic efficiency. In summary, UniBiomed represents a novel breakthrough in biomedical AI, unlocking powerful grounded interpretation capabilities for more accurate and efficient biomedical image analysis.
Diffusion-based Virtual Staining from Polarimetric Mueller Matrix Imaging
Mar 03, 2025


Abstract:Polarization, as a new optical imaging tool, has been explored to assist in the diagnosis of pathology. Moreover, converting the polarimetric Mueller Matrix (MM) to standardized stained images becomes a promising approach to help pathologists interpret the results. However, existing methods for polarization-based virtual staining are still in the early stage, and the diffusion-based model, which has shown great potential in enhancing the fidelity of the generated images, has not been studied yet. In this paper, a Regulated Bridge Diffusion Model (RBDM) for polarization-based virtual staining is proposed. RBDM utilizes the bidirectional bridge diffusion process to learn the mapping from polarization images to other modalities such as H\&E and fluorescence. And to demonstrate the effectiveness of our model, we conduct the experiment on our manually collected dataset, which consists of 18,000 paired polarization, fluorescence and H\&E images, due to the unavailability of the public dataset. The experiment results show that our model greatly outperforms other benchmark methods. Our dataset and code will be released upon acceptance.
FreeTumor: Large-Scale Generative Tumor Synthesis in Computed Tomography Images for Improving Tumor Recognition
Feb 23, 2025Abstract:Tumor is a leading cause of death worldwide, with an estimated 10 million deaths attributed to tumor-related diseases every year. AI-driven tumor recognition unlocks new possibilities for more precise and intelligent tumor screening and diagnosis. However, the progress is heavily hampered by the scarcity of annotated datasets, which demands extensive annotation efforts by radiologists. To tackle this challenge, we introduce FreeTumor, an innovative Generative AI (GAI) framework to enable large-scale tumor synthesis for mitigating data scarcity. Specifically, FreeTumor effectively leverages a combination of limited labeled data and large-scale unlabeled data for tumor synthesis training. Unleashing the power of large-scale data, FreeTumor is capable of synthesizing a large number of realistic tumors on images for augmenting training datasets. To this end, we create the largest training dataset for tumor synthesis and recognition by curating 161,310 publicly available Computed Tomography (CT) volumes from 33 sources, with only 2.3% containing annotated tumors. To validate the fidelity of synthetic tumors, we engaged 13 board-certified radiologists in a Visual Turing Test to discern between synthetic and real tumors. Rigorous clinician evaluation validates the high quality of our synthetic tumors, as they achieved only 51.1% sensitivity and 60.8% accuracy in distinguishing our synthetic tumors from real ones. Through high-quality tumor synthesis, FreeTumor scales up the recognition training datasets by over 40 times, showcasing a notable superiority over state-of-the-art AI methods including various synthesis methods and foundation models. These findings indicate promising prospects of FreeTumor in clinical applications, potentially advancing tumor treatments and improving the survival rates of patients.
MG-3D: Multi-Grained Knowledge-Enhanced 3D Medical Vision-Language Pre-training
Dec 08, 2024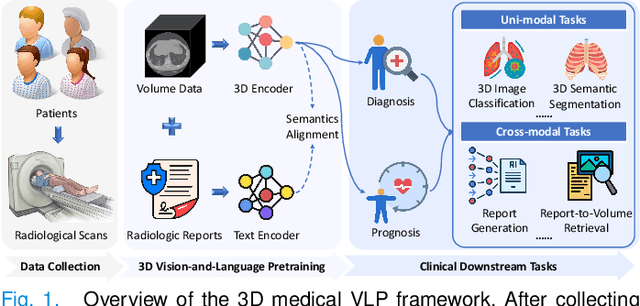

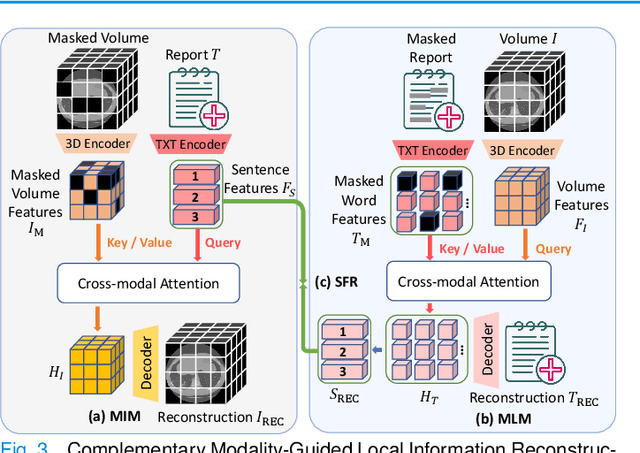

Abstract:3D medical image analysis is pivotal in numerous clinical applications. However, the scarcity of labeled data and limited generalization capabilities hinder the advancement of AI-empowered models. Radiology reports are easily accessible and can serve as weakly-supervised signals. However, large-scale vision-language pre-training (VLP) remains underexplored in 3D medical image analysis. Specifically, the insufficient investigation into multi-grained radiology semantics and their correlations across patients leads to underutilization of large-scale volume-report data. Considering intra-patient cross-modal semantic consistency and inter-patient semantic correlations, we propose a multi-task VLP method, MG-3D, pre-trained on large-scale data (47.1K), addressing the challenges by the following two aspects: 1) Establishing the correspondence between volume semantics and multi-grained medical knowledge of each patient with cross-modal global alignment and complementary modality-guided local reconstruction, ensuring intra-patient features of different modalities cohesively represent the same semantic content; 2) Correlating inter-patient visual semantics based on fine-grained report correlations across patients, and keeping sensitivity to global individual differences via contrastive learning, enhancing the discriminative feature representation. Furthermore, we delve into the scaling law to explore potential performance improvements. Comprehensive evaluations across nine uni- and cross-modal clinical tasks are carried out to assess model efficacy. Extensive experiments on both internal and external datasets demonstrate the superior transferability, scalability, and generalization of MG-3D, showcasing its potential in advancing feature representation for 3D medical image analysis. Code will be available: https://github.com/Xuefeng-Ni/MG-3D.
Touchstone Benchmark: Are We on the Right Way for Evaluating AI Algorithms for Medical Segmentation?
Nov 06, 2024



Abstract:How can we test AI performance? This question seems trivial, but it isn't. Standard benchmarks often have problems such as in-distribution and small-size test sets, oversimplified metrics, unfair comparisons, and short-term outcome pressure. As a consequence, good performance on standard benchmarks does not guarantee success in real-world scenarios. To address these problems, we present Touchstone, a large-scale collaborative segmentation benchmark of 9 types of abdominal organs. This benchmark is based on 5,195 training CT scans from 76 hospitals around the world and 5,903 testing CT scans from 11 additional hospitals. This diverse test set enhances the statistical significance of benchmark results and rigorously evaluates AI algorithms across various out-of-distribution scenarios. We invited 14 inventors of 19 AI algorithms to train their algorithms, while our team, as a third party, independently evaluated these algorithms on three test sets. In addition, we also evaluated pre-existing AI frameworks--which, differing from algorithms, are more flexible and can support different algorithms--including MONAI from NVIDIA, nnU-Net from DKFZ, and numerous other open-source frameworks. We are committed to expanding this benchmark to encourage more innovation of AI algorithms for the medical domain.
See it, Think it, Sorted: Large Multimodal Models are Few-shot Time Series Anomaly Analyzers
Nov 04, 2024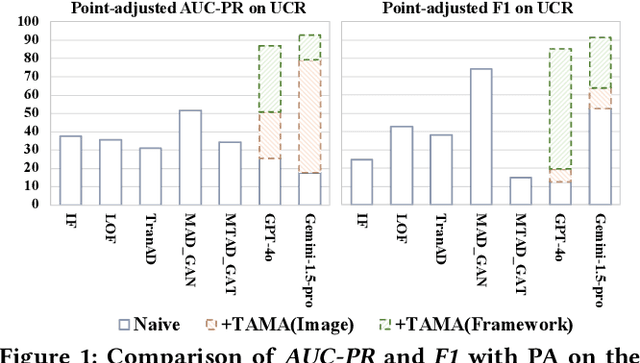

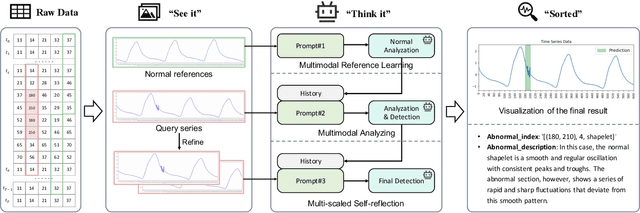

Abstract:Time series anomaly detection (TSAD) is becoming increasingly vital due to the rapid growth of time series data across various sectors. Anomalies in web service data, for example, can signal critical incidents such as system failures or server malfunctions, necessitating timely detection and response. However, most existing TSAD methodologies rely heavily on manual feature engineering or require extensive labeled training data, while also offering limited interpretability. To address these challenges, we introduce a pioneering framework called the Time Series Anomaly Multimodal Analyzer (TAMA), which leverages the power of Large Multimodal Models (LMMs) to enhance both the detection and interpretation of anomalies in time series data. By converting time series into visual formats that LMMs can efficiently process, TAMA leverages few-shot in-context learning capabilities to reduce dependence on extensive labeled datasets. Our methodology is validated through rigorous experimentation on multiple real-world datasets, where TAMA consistently outperforms state-of-the-art methods in TSAD tasks. Additionally, TAMA provides rich, natural language-based semantic analysis, offering deeper insights into the nature of detected anomalies. Furthermore, we contribute one of the first open-source datasets that includes anomaly detection labels, anomaly type labels, and contextual description, facilitating broader exploration and advancement within this critical field. Ultimately, TAMA not only excels in anomaly detection but also provides a comprehensive approach for understanding the underlying causes of anomalies, pushing TSAD forward through innovative methodologies and insights.
Large-Scale 3D Medical Image Pre-training with Geometric Context Priors
Oct 13, 2024Abstract:The scarcity of annotations poses a significant challenge in medical image analysis. Large-scale pre-training has emerged as a promising label-efficient solution, owing to the utilization of large-scale data, large models, and advanced pre-training techniques. However, its development in medical images remains underexplored. The primary challenge lies in harnessing large-scale unlabeled data and learning high-level semantics without annotations. We observe that 3D medical images exhibit consistent geometric context, i.e., consistent geometric relations between different organs, which leads to a promising way for learning consistent representations. Motivated by this, we introduce a simple-yet-effective Volume Contrast (VoCo) framework to leverage geometric context priors for self-supervision. Given an input volume, we extract base crops from different regions to construct positive and negative pairs for contrastive learning. Then we predict the contextual position of a random crop by contrasting its similarity to the base crops. In this way, VoCo encodes the inherent geometric context into model representations, facilitating high-level semantic learning without annotations. Specifically, we (1) introduce the largest medical pre-training dataset PreCT-160K; (2) investigate scaling laws and propose guidelines for tailoring different model sizes to various medical tasks; (3) build a benchmark encompassing 48 medical tasks. Extensive experiments highlight the superiority of VoCo. Codes at https://github.com/Luffy03/Large-Scale-Medical.
Unleashing the Denoising Capability of Diffusion Prior for Solving Inverse Problems
Jun 11, 2024Abstract:The recent emergence of diffusion models has significantly advanced the precision of learnable priors, presenting innovative avenues for addressing inverse problems. Since inverse problems inherently entail maximum a posteriori estimation, previous works have endeavored to integrate diffusion priors into the optimization frameworks. However, prevailing optimization-based inverse algorithms primarily exploit the prior information within the diffusion models while neglecting their denoising capability. To bridge this gap, this work leverages the diffusion process to reframe noisy inverse problems as a two-variable constrained optimization task by introducing an auxiliary optimization variable. By employing gradient truncation, the projection gradient descent method is efficiently utilized to solve the corresponding optimization problem. The proposed algorithm, termed ProjDiff, effectively harnesses the prior information and the denoising capability of a pre-trained diffusion model within the optimization framework. Extensive experiments on the image restoration tasks and source separation and partial generation tasks demonstrate that ProjDiff exhibits superior performance across various linear and nonlinear inverse problems, highlighting its potential for practical applications. Code is available at https://github.com/weigerzan/ProjDiff/.
FreeTumor: Advance Tumor Segmentation via Large-Scale Tumor Synthesis
Jun 03, 2024



Abstract:AI-driven tumor analysis has garnered increasing attention in healthcare. However, its progress is significantly hindered by the lack of annotated tumor cases, which requires radiologists to invest a lot of effort in collecting and annotation. In this paper, we introduce a highly practical solution for robust tumor synthesis and segmentation, termed FreeTumor, which refers to annotation-free synthetic tumors and our desire to free patients that suffering from tumors. Instead of pursuing sophisticated technical synthesis modules, we aim to design a simple yet effective tumor synthesis paradigm to unleash the power of large-scale data. Specifically, FreeTumor advances existing methods mainly from three aspects: (1) Existing methods only leverage small-scale labeled data for synthesis training, which limits their ability to generalize well on unseen data from different sources. To this end, we introduce the adversarial training strategy to leverage large-scale and diversified unlabeled data in synthesis training, significantly improving tumor synthesis. (2) Existing methods largely ignored the negative impact of low-quality synthetic tumors in segmentation training. Thus, we employ an adversarial-based discriminator to automatically filter out the low-quality synthetic tumors, which effectively alleviates their negative impact. (3) Existing methods only used hundreds of cases in tumor segmentation. In FreeTumor, we investigate the data scaling law in tumor segmentation by scaling up the dataset to 11k cases. Extensive experiments demonstrate the superiority of FreeTumor, e.g., on three tumor segmentation benchmarks, average $+8.9\%$ DSC over the baseline that only using real tumors and $+6.6\%$ DSC over the state-of-the-art tumor synthesis method. Code will be available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge