Qiong Wang
SAGS: Self-Adaptive Alias-Free Gaussian Splatting for Dynamic Surgical Endoscopic Reconstruction
Oct 31, 2025Abstract:Surgical reconstruction of dynamic tissues from endoscopic videos is a crucial technology in robot-assisted surgery. The development of Neural Radiance Fields (NeRFs) has greatly advanced deformable tissue reconstruction, achieving high-quality results from video and image sequences. However, reconstructing deformable endoscopic scenes remains challenging due to aliasing and artifacts caused by tissue movement, which can significantly degrade visualization quality. The introduction of 3D Gaussian Splatting (3DGS) has improved reconstruction efficiency by enabling a faster rendering pipeline. Nevertheless, existing 3DGS methods often prioritize rendering speed while neglecting these critical issues. To address these challenges, we propose SAGS, a self-adaptive alias-free Gaussian splatting framework. We introduce an attention-driven, dynamically weighted 4D deformation decoder, leveraging 3D smoothing filters and 2D Mip filters to mitigate artifacts in deformable tissue reconstruction and better capture the fine details of tissue movement. Experimental results on two public benchmarks, EndoNeRF and SCARED, demonstrate that our method achieves superior performance in all metrics of PSNR, SSIM, and LPIPS compared to the state of the art while also delivering better visualization quality.
DR-SAC: Distributionally Robust Soft Actor-Critic for Reinforcement Learning under Uncertainty
Jun 14, 2025Abstract:Deep reinforcement learning (RL) has achieved significant success, yet its application in real-world scenarios is often hindered by a lack of robustness to environmental uncertainties. To solve this challenge, some robust RL algorithms have been proposed, but most are limited to tabular settings. In this work, we propose Distributionally Robust Soft Actor-Critic (DR-SAC), a novel algorithm designed to enhance the robustness of the state-of-the-art Soft Actor-Critic (SAC) algorithm. DR-SAC aims to maximize the expected value with entropy against the worst possible transition model lying in an uncertainty set. A distributionally robust version of the soft policy iteration is derived with a convergence guarantee. For settings where nominal distributions are unknown, such as offline RL, a generative modeling approach is proposed to estimate the required nominal distributions from data. Furthermore, experimental results on a range of continuous control benchmark tasks demonstrate our algorithm achieves up to $9.8$ times the average reward of the SAC baseline under common perturbations. Additionally, compared with existing robust reinforcement learning algorithms, DR-SAC significantly improves computing efficiency and applicability to large-scale problems.
MG-3D: Multi-Grained Knowledge-Enhanced 3D Medical Vision-Language Pre-training
Dec 08, 2024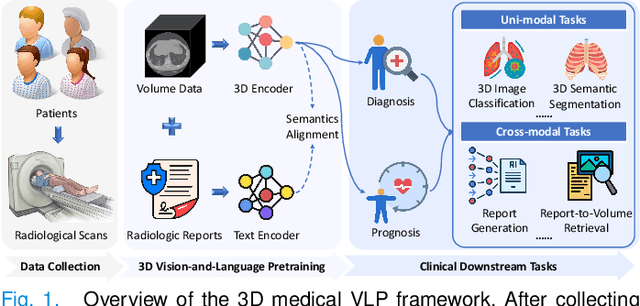

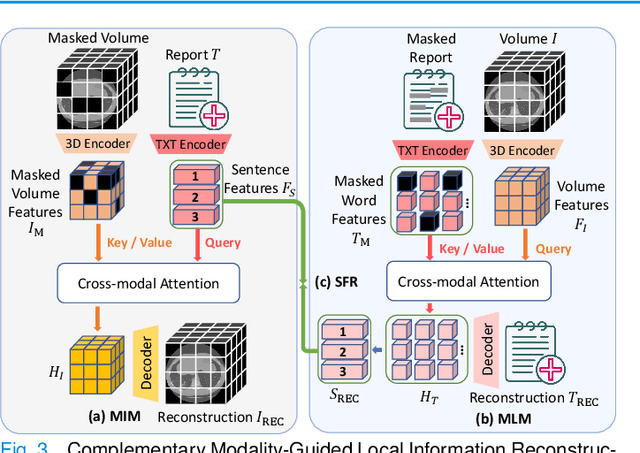

Abstract:3D medical image analysis is pivotal in numerous clinical applications. However, the scarcity of labeled data and limited generalization capabilities hinder the advancement of AI-empowered models. Radiology reports are easily accessible and can serve as weakly-supervised signals. However, large-scale vision-language pre-training (VLP) remains underexplored in 3D medical image analysis. Specifically, the insufficient investigation into multi-grained radiology semantics and their correlations across patients leads to underutilization of large-scale volume-report data. Considering intra-patient cross-modal semantic consistency and inter-patient semantic correlations, we propose a multi-task VLP method, MG-3D, pre-trained on large-scale data (47.1K), addressing the challenges by the following two aspects: 1) Establishing the correspondence between volume semantics and multi-grained medical knowledge of each patient with cross-modal global alignment and complementary modality-guided local reconstruction, ensuring intra-patient features of different modalities cohesively represent the same semantic content; 2) Correlating inter-patient visual semantics based on fine-grained report correlations across patients, and keeping sensitivity to global individual differences via contrastive learning, enhancing the discriminative feature representation. Furthermore, we delve into the scaling law to explore potential performance improvements. Comprehensive evaluations across nine uni- and cross-modal clinical tasks are carried out to assess model efficacy. Extensive experiments on both internal and external datasets demonstrate the superior transferability, scalability, and generalization of MG-3D, showcasing its potential in advancing feature representation for 3D medical image analysis. Code will be available: https://github.com/Xuefeng-Ni/MG-3D.
Diff-VPS: Video Polyp Segmentation via a Multi-task Diffusion Network with Adversarial Temporal Reasoning
Sep 11, 2024Abstract:Diffusion Probabilistic Models have recently attracted significant attention in the community of computer vision due to their outstanding performance. However, while a substantial amount of diffusion-based research has focused on generative tasks, no work introduces diffusion models to advance the results of polyp segmentation in videos, which is frequently challenged by polyps' high camouflage and redundant temporal cues.In this paper, we present a novel diffusion-based network for video polyp segmentation task, dubbed as Diff-VPS. We incorporate multi-task supervision into diffusion models to promote the discrimination of diffusion models on pixel-by-pixel segmentation. This integrates the contextual high-level information achieved by the joint classification and detection tasks. To explore the temporal dependency, Temporal Reasoning Module (TRM) is devised via reasoning and reconstructing the target frame from the previous frames. We further equip TRM with a generative adversarial self-supervised strategy to produce more realistic frames and thus capture better dynamic cues. Extensive experiments are conducted on SUN-SEG, and the results indicate that our proposed Diff-VPS significantly achieves state-of-the-art performance. Code is available at https://github.com/lydia-yllu/Diff-VPS.
Timeline and Boundary Guided Diffusion Network for Video Shadow Detection
Aug 21, 2024



Abstract:Video Shadow Detection (VSD) aims to detect the shadow masks with frame sequence. Existing works suffer from inefficient temporal learning. Moreover, few works address the VSD problem by considering the characteristic (i.e., boundary) of shadow. Motivated by this, we propose a Timeline and Boundary Guided Diffusion (TBGDiff) network for VSD where we take account of the past-future temporal guidance and boundary information jointly. In detail, we design a Dual Scale Aggregation (DSA) module for better temporal understanding by rethinking the affinity of the long-term and short-term frames for the clipped video. Next, we introduce Shadow Boundary Aware Attention (SBAA) to utilize the edge contexts for capturing the characteristics of shadows. Moreover, we are the first to introduce the Diffusion model for VSD in which we explore a Space-Time Encoded Embedding (STEE) to inject the temporal guidance for Diffusion to conduct shadow detection. Benefiting from these designs, our model can not only capture the temporal information but also the shadow property. Extensive experiments show that the performance of our approach overtakes the state-of-the-art methods, verifying the effectiveness of our components. We release the codes, weights, and results at \url{https://github.com/haipengzhou856/TBGDiff}.
Surgformer: Surgical Transformer with Hierarchical Temporal Attention for Surgical Phase Recognition
Aug 07, 2024Abstract:Existing state-of-the-art methods for surgical phase recognition either rely on the extraction of spatial-temporal features at a short-range temporal resolution or adopt the sequential extraction of the spatial and temporal features across the entire temporal resolution. However, these methods have limitations in modeling spatial-temporal dependency and addressing spatial-temporal redundancy: 1) These methods fail to effectively model spatial-temporal dependency, due to the lack of long-range information or joint spatial-temporal modeling. 2) These methods utilize dense spatial features across the entire temporal resolution, resulting in significant spatial-temporal redundancy. In this paper, we propose the Surgical Transformer (Surgformer) to address the issues of spatial-temporal modeling and redundancy in an end-to-end manner, which employs divided spatial-temporal attention and takes a limited set of sparse frames as input. Moreover, we propose a novel Hierarchical Temporal Attention (HTA) to capture both global and local information within varied temporal resolutions from a target frame-centric perspective. Distinct from conventional temporal attention that primarily emphasizes dense long-range similarity, HTA not only captures long-term information but also considers local latent consistency among informative frames. HTA then employs pyramid feature aggregation to effectively utilize temporal information across diverse temporal resolutions, thereby enhancing the overall temporal representation. Extensive experiments on two challenging benchmark datasets verify that our proposed Surgformer performs favorably against the state-of-the-art methods. The code is released at https://github.com/isyangshu/Surgformer.
Advancing UWF-SLO Vessel Segmentation with Source-Free Active Domain Adaptation and a Novel Multi-Center Dataset
Jun 19, 2024Abstract:Accurate vessel segmentation in Ultra-Wide-Field Scanning Laser Ophthalmoscopy (UWF-SLO) images is crucial for diagnosing retinal diseases. Although recent techniques have shown encouraging outcomes in vessel segmentation, models trained on one medical dataset often underperform on others due to domain shifts. Meanwhile, manually labeling high-resolution UWF-SLO images is an extremely challenging, time-consuming and expensive task. In response, this study introduces a pioneering framework that leverages a patch-based active domain adaptation approach. By actively recommending a few valuable image patches by the devised Cascade Uncertainty-Predominance (CUP) selection strategy for labeling and model-finetuning, our method significantly improves the accuracy of UWF-SLO vessel segmentation across diverse medical centers. In addition, we annotate and construct the first Multi-center UWF-SLO Vessel Segmentation (MU-VS) dataset to promote this topic research, comprising data from multiple institutions. This dataset serves as a valuable resource for cross-center evaluation, verifying the effectiveness and robustness of our approach. Experimental results demonstrate that our approach surpasses existing domain adaptation and active learning methods, considerably reducing the gap between the Upper and Lower bounds with minimal annotations, highlighting our method's practical clinical value. We will release our dataset and code to facilitate relevant research: https://github.com/whq-xxh/SFADA-UWF-SLO.
Ultrasound Report Generation with Cross-Modality Feature Alignment via Unsupervised Guidance
Jun 02, 2024Abstract:Automatic report generation has arisen as a significant research area in computer-aided diagnosis, aiming to alleviate the burden on clinicians by generating reports automatically based on medical images. In this work, we propose a novel framework for automatic ultrasound report generation, leveraging a combination of unsupervised and supervised learning methods to aid the report generation process. Our framework incorporates unsupervised learning methods to extract potential knowledge from ultrasound text reports, serving as the prior information to guide the model in aligning visual and textual features, thereby addressing the challenge of feature discrepancy. Additionally, we design a global semantic comparison mechanism to enhance the performance of generating more comprehensive and accurate medical reports. To enable the implementation of ultrasound report generation, we constructed three large-scale ultrasound image-text datasets from different organs for training and validation purposes. Extensive evaluations with other state-of-the-art approaches exhibit its superior performance across all three datasets. Code and dataset are valuable at this link.
MiM: Mask in Mask Self-Supervised Pre-Training for 3D Medical Image Analysis
Apr 24, 2024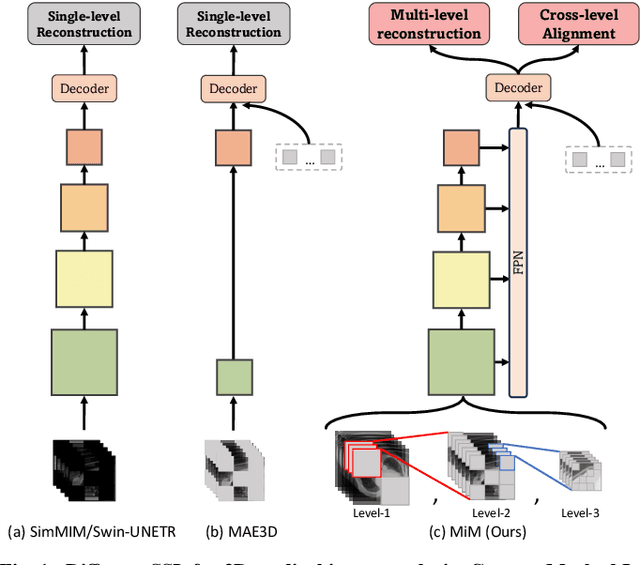
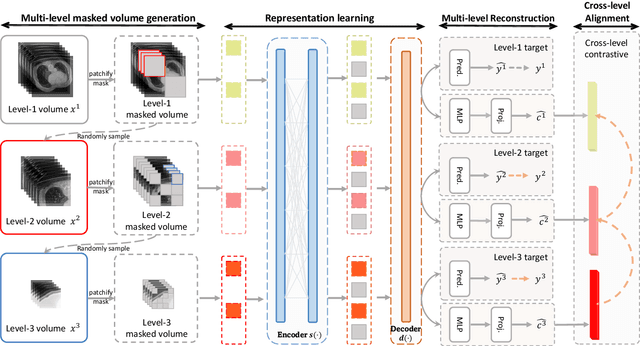
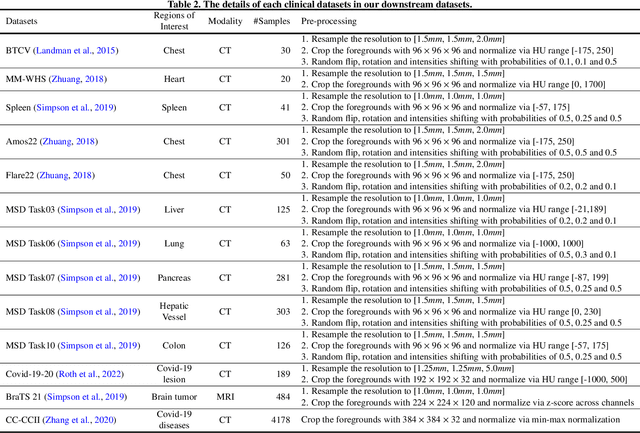
Abstract:The Vision Transformer (ViT) has demonstrated remarkable performance in Self-Supervised Learning (SSL) for 3D medical image analysis. Mask AutoEncoder (MAE) for feature pre-training can further unleash the potential of ViT on various medical vision tasks. However, due to large spatial sizes with much higher dimensions of 3D medical images, the lack of hierarchical design for MAE may hinder the performance of downstream tasks. In this paper, we propose a novel \textit{Mask in Mask (MiM)} pre-training framework for 3D medical images, which aims to advance MAE by learning discriminative representation from hierarchical visual tokens across varying scales. We introduce multiple levels of granularity for masked inputs from the volume, which are then reconstructed simultaneously ranging at both fine and coarse levels. Additionally, a cross-level alignment mechanism is applied to adjacent level volumes to enforce anatomical similarity hierarchically. Furthermore, we adopt a hybrid backbone to enhance the hierarchical representation learning efficiently during the pre-training. MiM was pre-trained on a large scale of available 3D volumetric images, \textit{i.e.,} Computed Tomography (CT) images containing various body parts. Extensive experiments on thirteen public datasets demonstrate the superiority of MiM over other SSL methods in organ/lesion/tumor segmentation and disease classification. We further scale up the MiM to large pre-training datasets with more than 10k volumes, showing that large-scale pre-training can further enhance the performance of downstream tasks. The improvement also concluded that the research community should pay more attention to the scale of the pre-training dataset towards the healthcare foundation model for 3D medical images.
Design as Desired: Utilizing Visual Question Answering for Multimodal Pre-training
Apr 08, 2024Abstract:Multimodal pre-training demonstrates its potential in the medical domain, which learns medical visual representations from paired medical reports. However, many pre-training tasks require extra annotations from clinicians, and most of them fail to explicitly guide the model to learn the desired features of different pathologies. To the best of our knowledge, we are the first to utilize Visual Question Answering (VQA) for multimodal pre-training to guide the framework focusing on targeted pathological features. In this work, we leverage descriptions in medical reports to design multi-granular question-answer pairs associated with different diseases, which assist the framework in pre-training without requiring extra annotations from experts. We also propose a novel pre-training framework with a quasi-textual feature transformer, a module designed to transform visual features into a quasi-textual space closer to the textual domain via a contrastive learning strategy. This narrows the vision-language gap and facilitates modality alignment. Our framework is applied to four downstream tasks: report generation, classification, segmentation, and detection across five datasets. Extensive experiments demonstrate the superiority of our framework compared to other state-of-the-art methods. Our code will be released upon acceptance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge