Mingxiang Wu
FreeTumor: Large-Scale Generative Tumor Synthesis in Computed Tomography Images for Improving Tumor Recognition
Feb 23, 2025Abstract:Tumor is a leading cause of death worldwide, with an estimated 10 million deaths attributed to tumor-related diseases every year. AI-driven tumor recognition unlocks new possibilities for more precise and intelligent tumor screening and diagnosis. However, the progress is heavily hampered by the scarcity of annotated datasets, which demands extensive annotation efforts by radiologists. To tackle this challenge, we introduce FreeTumor, an innovative Generative AI (GAI) framework to enable large-scale tumor synthesis for mitigating data scarcity. Specifically, FreeTumor effectively leverages a combination of limited labeled data and large-scale unlabeled data for tumor synthesis training. Unleashing the power of large-scale data, FreeTumor is capable of synthesizing a large number of realistic tumors on images for augmenting training datasets. To this end, we create the largest training dataset for tumor synthesis and recognition by curating 161,310 publicly available Computed Tomography (CT) volumes from 33 sources, with only 2.3% containing annotated tumors. To validate the fidelity of synthetic tumors, we engaged 13 board-certified radiologists in a Visual Turing Test to discern between synthetic and real tumors. Rigorous clinician evaluation validates the high quality of our synthetic tumors, as they achieved only 51.1% sensitivity and 60.8% accuracy in distinguishing our synthetic tumors from real ones. Through high-quality tumor synthesis, FreeTumor scales up the recognition training datasets by over 40 times, showcasing a notable superiority over state-of-the-art AI methods including various synthesis methods and foundation models. These findings indicate promising prospects of FreeTumor in clinical applications, potentially advancing tumor treatments and improving the survival rates of patients.
MG-3D: Multi-Grained Knowledge-Enhanced 3D Medical Vision-Language Pre-training
Dec 08, 2024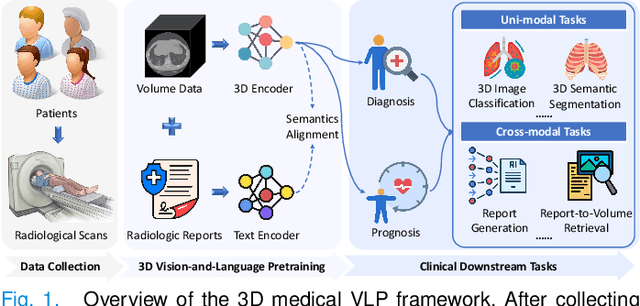

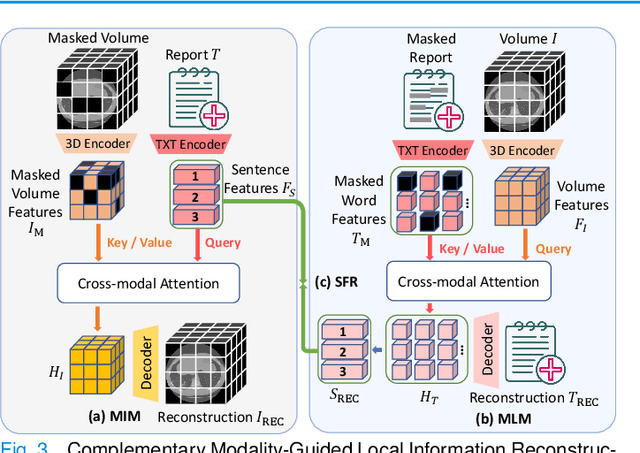

Abstract:3D medical image analysis is pivotal in numerous clinical applications. However, the scarcity of labeled data and limited generalization capabilities hinder the advancement of AI-empowered models. Radiology reports are easily accessible and can serve as weakly-supervised signals. However, large-scale vision-language pre-training (VLP) remains underexplored in 3D medical image analysis. Specifically, the insufficient investigation into multi-grained radiology semantics and their correlations across patients leads to underutilization of large-scale volume-report data. Considering intra-patient cross-modal semantic consistency and inter-patient semantic correlations, we propose a multi-task VLP method, MG-3D, pre-trained on large-scale data (47.1K), addressing the challenges by the following two aspects: 1) Establishing the correspondence between volume semantics and multi-grained medical knowledge of each patient with cross-modal global alignment and complementary modality-guided local reconstruction, ensuring intra-patient features of different modalities cohesively represent the same semantic content; 2) Correlating inter-patient visual semantics based on fine-grained report correlations across patients, and keeping sensitivity to global individual differences via contrastive learning, enhancing the discriminative feature representation. Furthermore, we delve into the scaling law to explore potential performance improvements. Comprehensive evaluations across nine uni- and cross-modal clinical tasks are carried out to assess model efficacy. Extensive experiments on both internal and external datasets demonstrate the superior transferability, scalability, and generalization of MG-3D, showcasing its potential in advancing feature representation for 3D medical image analysis. Code will be available: https://github.com/Xuefeng-Ni/MG-3D.
Charting the Path Forward: CT Image Quality Assessment -- An In-Depth Review
Apr 30, 2024



Abstract:Computed Tomography (CT) is a frequently utilized imaging technology that is employed in the clinical diagnosis of many disorders. However, clinical diagnosis, data storage, and management are posed huge challenges by a huge volume of non-homogeneous CT data in terms of imaging quality. As a result, the quality assessment of CT images is a crucial problem that demands consideration. The history, advancements in research, and current developments in CT image quality assessment (IQA) are examined in this paper. In this review, we collected and researched more than 500 CT-IQA publications published before August 2023. And we provide the visualization analysis of keywords and co-citations in the knowledge graph of these papers. Prospects and obstacles for the continued development of CT-IQA are also covered. At present, significant research branches in the CT-IQA domain include Phantom study, Artificial intelligence deep-learning reconstruction algorithm, Dose reduction opportunity, and Virtual monoenergetic reconstruction. Artificial intelligence (AI)-based CT-IQA also becomes a trend. It increases the accuracy of the CT scanning apparatus, amplifies the impact of the CT system reconstruction algorithm, and creates an effective algorithm for post-processing CT images. AI-based medical IQA offers excellent application opportunities in clinical work. AI can provide uniform quality assessment criteria and more comprehensive guidance amongst various healthcare facilities, and encourage them to identify one another's images. It will help lower the number of unnecessary tests and associated costs, and enhance the quality of medical imaging and assessment efficiency.
Learning Agreement from Multi-source Annotations for Medical Image Segmentation
Apr 02, 2023



Abstract:In medical image analysis, it is typical to merge multiple independent annotations as ground truth to mitigate the bias caused by individual annotation preference. However, arbitrating the final annotation is not always effective because new biases might be produced during the process, especially when there are significant variations among annotations. This paper proposes a novel Uncertainty-guided Multi-source Annotation Network (UMA-Net) to learn medical image segmentation directly from multiple annotations. UMA-Net consists of a UNet with two quality-specific predictors, an Annotation Uncertainty Estimation Module (AUEM) and a Quality Assessment Module (QAM). Specifically, AUEM estimates pixel-wise uncertainty maps of each annotation and encourages them to reach an agreement on reliable pixels/voxels. The uncertainty maps then guide the UNet to learn from the reliable pixels/voxels by weighting the segmentation loss. QAM grades the uncertainty maps into high-quality or low-quality groups based on assessment scores. The UNet is further implemented to contain a high-quality learning head (H-head) and a low-quality learning head (L-head). H-head purely learns with high-quality uncertainty maps to avoid error accumulation and keeps strong prediction ability, while L-head leverages the low-quality uncertainty maps to assist the backbone to learn maximum representation knowledge. UNet with H-head will be reserved during the inference stage, and the rest of the modules can be removed freely for computational efficiency. We conduct extensive experiments on an unsupervised 3D segmentation task and a supervised 2D segmentation task, respectively. The results show that our proposed UMA-Net outperforms state-of-the-art approaches, demonstrating its generality and effectiveness.
Rethinking annotation granularity for overcoming deep shortcut learning: A retrospective study on chest radiographs
Apr 21, 2021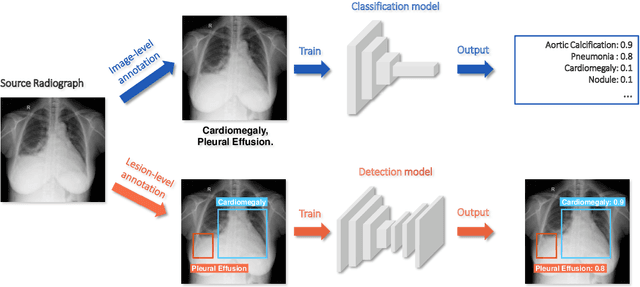
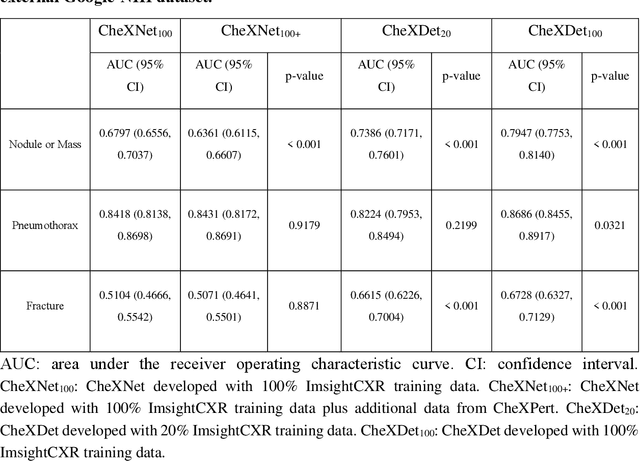
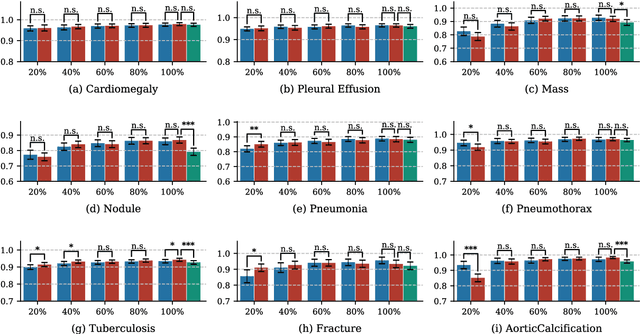
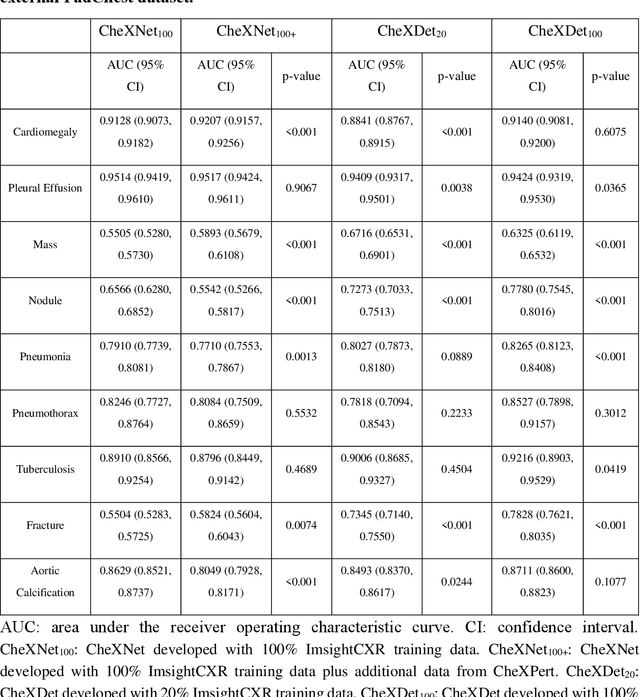
Abstract:Deep learning has demonstrated radiograph screening performances that are comparable or superior to radiologists. However, recent studies show that deep models for thoracic disease classification usually show degraded performance when applied to external data. Such phenomena can be categorized into shortcut learning, where the deep models learn unintended decision rules that can fit the identically distributed training and test set but fail to generalize to other distributions. A natural way to alleviate this defect is explicitly indicating the lesions and focusing the model on learning the intended features. In this paper, we conduct extensive retrospective experiments to compare a popular thoracic disease classification model, CheXNet, and a thoracic lesion detection model, CheXDet. We first showed that the two models achieved similar image-level classification performance on the internal test set with no significant differences under many scenarios. Meanwhile, we found incorporating external training data even led to performance degradation for CheXNet. Then, we compared the models' internal performance on the lesion localization task and showed that CheXDet achieved significantly better performance than CheXNet even when given 80% less training data. By further visualizing the models' decision-making regions, we revealed that CheXNet learned patterns other than the target lesions, demonstrating its shortcut learning defect. Moreover, CheXDet achieved significantly better external performance than CheXNet on both the image-level classification task and the lesion localization task. Our findings suggest improving annotation granularity for training deep learning systems as a promising way to elevate future deep learning-based diagnosis systems for clinical usage.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge