Xuefeng Ni
FreeTumor: Large-Scale Generative Tumor Synthesis in Computed Tomography Images for Improving Tumor Recognition
Feb 23, 2025Abstract:Tumor is a leading cause of death worldwide, with an estimated 10 million deaths attributed to tumor-related diseases every year. AI-driven tumor recognition unlocks new possibilities for more precise and intelligent tumor screening and diagnosis. However, the progress is heavily hampered by the scarcity of annotated datasets, which demands extensive annotation efforts by radiologists. To tackle this challenge, we introduce FreeTumor, an innovative Generative AI (GAI) framework to enable large-scale tumor synthesis for mitigating data scarcity. Specifically, FreeTumor effectively leverages a combination of limited labeled data and large-scale unlabeled data for tumor synthesis training. Unleashing the power of large-scale data, FreeTumor is capable of synthesizing a large number of realistic tumors on images for augmenting training datasets. To this end, we create the largest training dataset for tumor synthesis and recognition by curating 161,310 publicly available Computed Tomography (CT) volumes from 33 sources, with only 2.3% containing annotated tumors. To validate the fidelity of synthetic tumors, we engaged 13 board-certified radiologists in a Visual Turing Test to discern between synthetic and real tumors. Rigorous clinician evaluation validates the high quality of our synthetic tumors, as they achieved only 51.1% sensitivity and 60.8% accuracy in distinguishing our synthetic tumors from real ones. Through high-quality tumor synthesis, FreeTumor scales up the recognition training datasets by over 40 times, showcasing a notable superiority over state-of-the-art AI methods including various synthesis methods and foundation models. These findings indicate promising prospects of FreeTumor in clinical applications, potentially advancing tumor treatments and improving the survival rates of patients.
MG-3D: Multi-Grained Knowledge-Enhanced 3D Medical Vision-Language Pre-training
Dec 08, 2024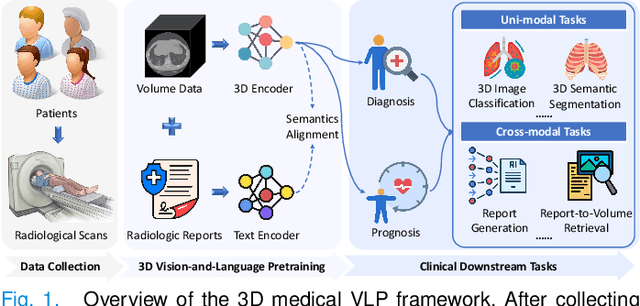

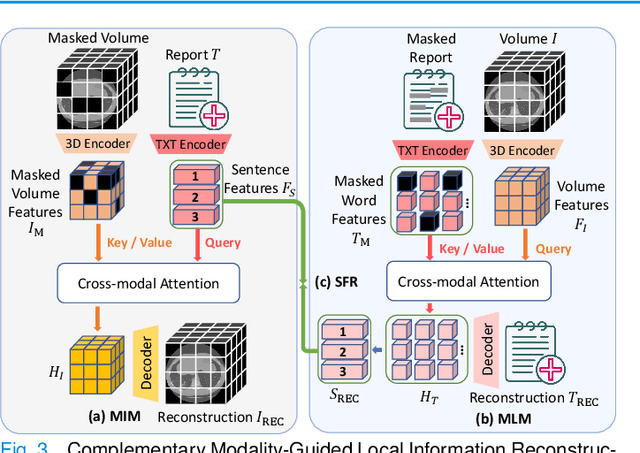

Abstract:3D medical image analysis is pivotal in numerous clinical applications. However, the scarcity of labeled data and limited generalization capabilities hinder the advancement of AI-empowered models. Radiology reports are easily accessible and can serve as weakly-supervised signals. However, large-scale vision-language pre-training (VLP) remains underexplored in 3D medical image analysis. Specifically, the insufficient investigation into multi-grained radiology semantics and their correlations across patients leads to underutilization of large-scale volume-report data. Considering intra-patient cross-modal semantic consistency and inter-patient semantic correlations, we propose a multi-task VLP method, MG-3D, pre-trained on large-scale data (47.1K), addressing the challenges by the following two aspects: 1) Establishing the correspondence between volume semantics and multi-grained medical knowledge of each patient with cross-modal global alignment and complementary modality-guided local reconstruction, ensuring intra-patient features of different modalities cohesively represent the same semantic content; 2) Correlating inter-patient visual semantics based on fine-grained report correlations across patients, and keeping sensitivity to global individual differences via contrastive learning, enhancing the discriminative feature representation. Furthermore, we delve into the scaling law to explore potential performance improvements. Comprehensive evaluations across nine uni- and cross-modal clinical tasks are carried out to assess model efficacy. Extensive experiments on both internal and external datasets demonstrate the superior transferability, scalability, and generalization of MG-3D, showcasing its potential in advancing feature representation for 3D medical image analysis. Code will be available: https://github.com/Xuefeng-Ni/MG-3D.
FreeTumor: Advance Tumor Segmentation via Large-Scale Tumor Synthesis
Jun 03, 2024



Abstract:AI-driven tumor analysis has garnered increasing attention in healthcare. However, its progress is significantly hindered by the lack of annotated tumor cases, which requires radiologists to invest a lot of effort in collecting and annotation. In this paper, we introduce a highly practical solution for robust tumor synthesis and segmentation, termed FreeTumor, which refers to annotation-free synthetic tumors and our desire to free patients that suffering from tumors. Instead of pursuing sophisticated technical synthesis modules, we aim to design a simple yet effective tumor synthesis paradigm to unleash the power of large-scale data. Specifically, FreeTumor advances existing methods mainly from three aspects: (1) Existing methods only leverage small-scale labeled data for synthesis training, which limits their ability to generalize well on unseen data from different sources. To this end, we introduce the adversarial training strategy to leverage large-scale and diversified unlabeled data in synthesis training, significantly improving tumor synthesis. (2) Existing methods largely ignored the negative impact of low-quality synthetic tumors in segmentation training. Thus, we employ an adversarial-based discriminator to automatically filter out the low-quality synthetic tumors, which effectively alleviates their negative impact. (3) Existing methods only used hundreds of cases in tumor segmentation. In FreeTumor, we investigate the data scaling law in tumor segmentation by scaling up the dataset to 11k cases. Extensive experiments demonstrate the superiority of FreeTumor, e.g., on three tumor segmentation benchmarks, average $+8.9\%$ DSC over the baseline that only using real tumors and $+6.6\%$ DSC over the state-of-the-art tumor synthesis method. Code will be available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge