Yifeng Wang
A Dual-Branch Local-Global Framework for Cross-Resolution Land Cover Mapping
Dec 23, 2025Abstract:Cross-resolution land cover mapping aims to produce high-resolution semantic predictions from coarse or low-resolution supervision, yet the severe resolution mismatch makes effective learning highly challenging. Existing weakly supervised approaches often struggle to align fine-grained spatial structures with coarse labels, leading to noisy supervision and degraded mapping accuracy. To tackle this problem, we propose DDTM, a dual-branch weakly supervised framework that explicitly decouples local semantic refinement from global contextual reasoning. Specifically, DDTM introduces a diffusion-based branch to progressively refine fine-scale local semantics under coarse supervision, while a transformer-based branch enforces long-range contextual consistency across large spatial extents. In addition, we design a pseudo-label confidence evaluation module to mitigate noise induced by cross-resolution inconsistencies and to selectively exploit reliable supervisory signals. Extensive experiments demonstrate that DDTM establishes a new state-of-the-art on the Chesapeake Bay benchmark, achieving 66.52\% mIoU and substantially outperforming prior weakly supervised methods. The code is available at https://github.com/gpgpgp123/DDTM.
A Semantically Enhanced Generative Foundation Model Improves Pathological Image Synthesis
Dec 16, 2025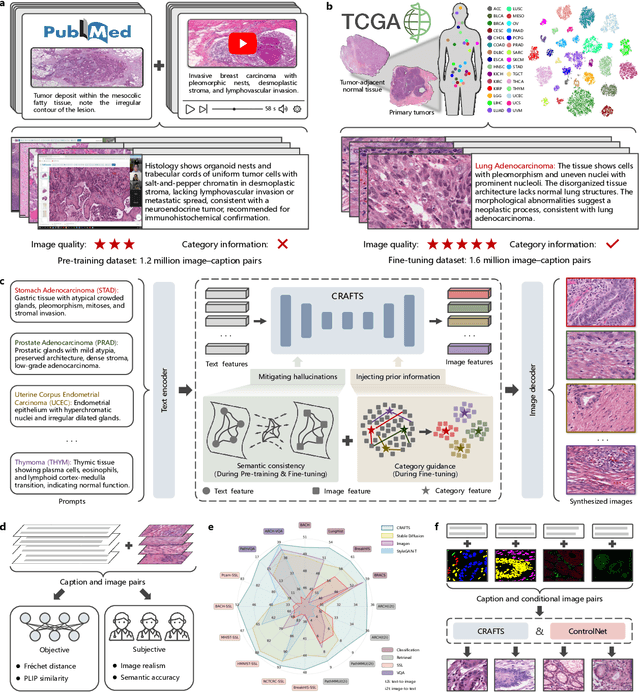
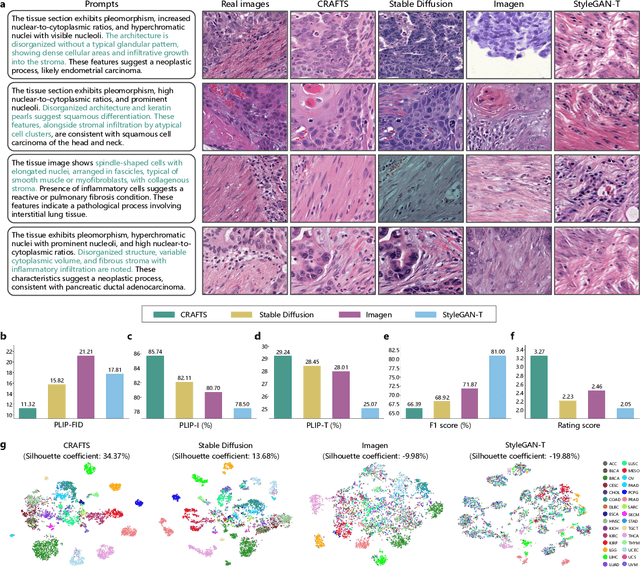
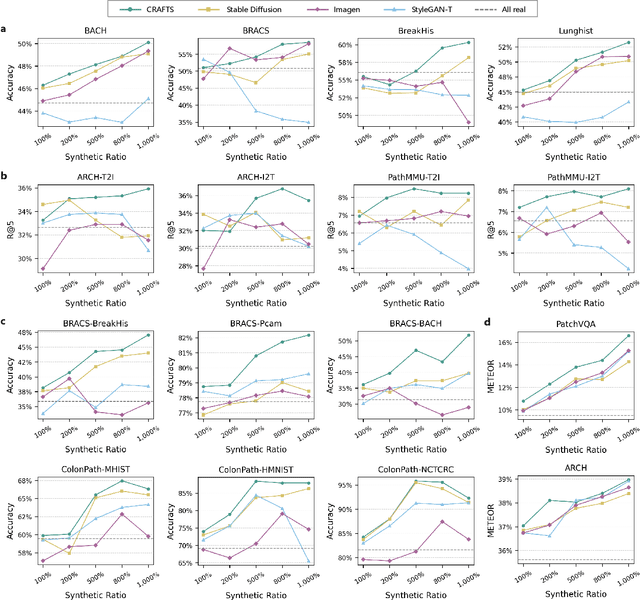
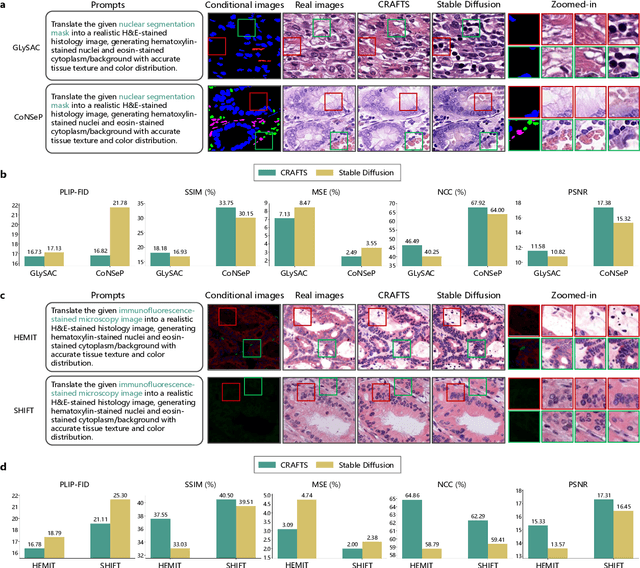
Abstract:The development of clinical-grade artificial intelligence in pathology is limited by the scarcity of diverse, high-quality annotated datasets. Generative models offer a potential solution but suffer from semantic instability and morphological hallucinations that compromise diagnostic reliability. To address this challenge, we introduce a Correlation-Regulated Alignment Framework for Tissue Synthesis (CRAFTS), the first generative foundation model for pathology-specific text-to-image synthesis. By leveraging a dual-stage training strategy on approximately 2.8 million image-caption pairs, CRAFTS incorporates a novel alignment mechanism that suppresses semantic drift to ensure biological accuracy. This model generates diverse pathological images spanning 30 cancer types, with quality rigorously validated by objective metrics and pathologist evaluations. Furthermore, CRAFTS-augmented datasets enhance the performance across various clinical tasks, including classification, cross-modal retrieval, self-supervised learning, and visual question answering. In addition, coupling CRAFTS with ControlNet enables precise control over tissue architecture from inputs such as nuclear segmentation masks and fluorescence images. By overcoming the critical barriers of data scarcity and privacy concerns, CRAFTS provides a limitless source of diverse, annotated histology data, effectively unlocking the creation of robust diagnostic tools for rare and complex cancer phenotypes.
RubikSQL: Lifelong Learning Agentic Knowledge Base as an Industrial NL2SQL System
Aug 25, 2025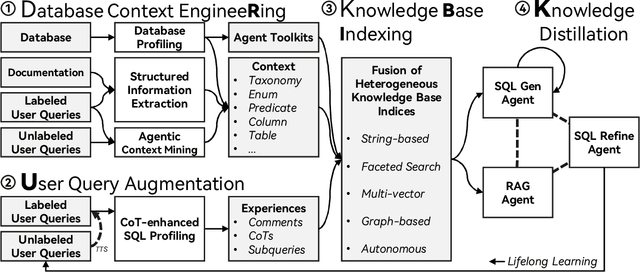
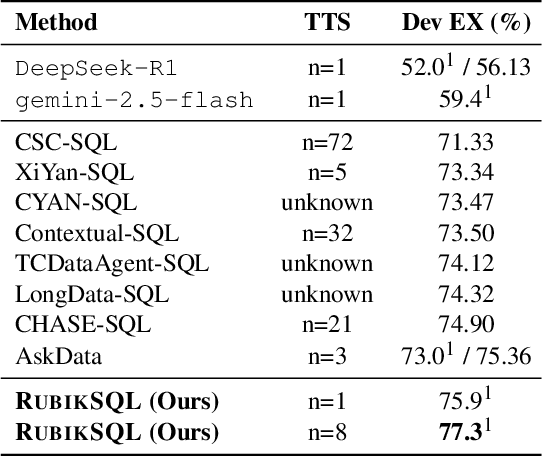
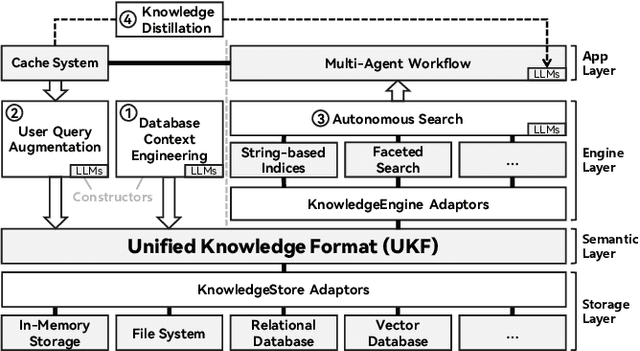
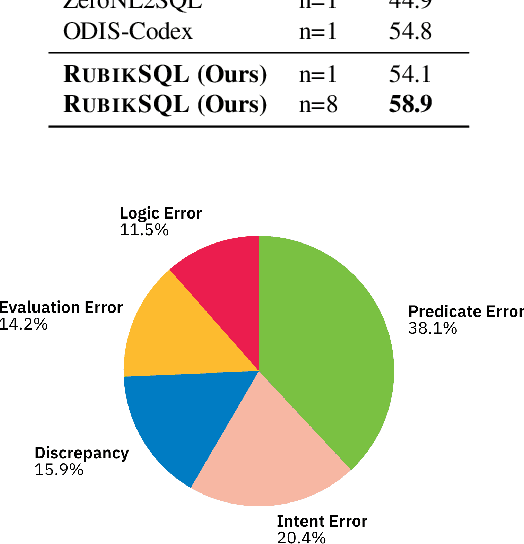
Abstract:We present RubikSQL, a novel NL2SQL system designed to address key challenges in real-world enterprise-level NL2SQL, such as implicit intents and domain-specific terminology. RubikSQL frames NL2SQL as a lifelong learning task, demanding both Knowledge Base (KB) maintenance and SQL generation. RubikSQL systematically builds and refines its KB through techniques including database profiling, structured information extraction, agentic rule mining, and Chain-of-Thought (CoT)-enhanced SQL profiling. RubikSQL then employs a multi-agent workflow to leverage this curated KB, generating accurate SQLs. RubikSQL achieves SOTA performance on both the KaggleDBQA and BIRD Mini-Dev datasets. Finally, we release the RubikBench benchmark, a new benchmark specifically designed to capture vital traits of industrial NL2SQL scenarios, providing a valuable resource for future research.
The Four Color Theorem for Cell Instance Segmentation
Jun 11, 2025Abstract:Cell instance segmentation is critical to analyzing biomedical images, yet accurately distinguishing tightly touching cells remains a persistent challenge. Existing instance segmentation frameworks, including detection-based, contour-based, and distance mapping-based approaches, have made significant progress, but balancing model performance with computational efficiency remains an open problem. In this paper, we propose a novel cell instance segmentation method inspired by the four-color theorem. By conceptualizing cells as countries and tissues as oceans, we introduce a four-color encoding scheme that ensures adjacent instances receive distinct labels. This reformulation transforms instance segmentation into a constrained semantic segmentation problem with only four predicted classes, substantially simplifying the instance differentiation process. To solve the training instability caused by the non-uniqueness of four-color encoding, we design an asymptotic training strategy and encoding transformation method. Extensive experiments on various modes demonstrate our approach achieves state-of-the-art performance. The code is available at https://github.com/zhangye-zoe/FCIS.
OWL: Optimized Workforce Learning for General Multi-Agent Assistance in Real-World Task Automation
May 29, 2025Abstract:Large Language Model (LLM)-based multi-agent systems show promise for automating real-world tasks but struggle to transfer across domains due to their domain-specific nature. Current approaches face two critical shortcomings: they require complete architectural redesign and full retraining of all components when applied to new domains. We introduce Workforce, a hierarchical multi-agent framework that decouples strategic planning from specialized execution through a modular architecture comprising: (i) a domain-agnostic Planner for task decomposition, (ii) a Coordinator for subtask management, and (iii) specialized Workers with domain-specific tool-calling capabilities. This decoupling enables cross-domain transferability during both inference and training phases: During inference, Workforce seamlessly adapts to new domains by adding or modifying worker agents; For training, we introduce Optimized Workforce Learning (OWL), which improves generalization across domains by optimizing a domain-agnostic planner with reinforcement learning from real-world feedback. To validate our approach, we evaluate Workforce on the GAIA benchmark, covering various realistic, multi-domain agentic tasks. Experimental results demonstrate Workforce achieves open-source state-of-the-art performance (69.70%), outperforming commercial systems like OpenAI's Deep Research by 2.34%. More notably, our OWL-trained 32B model achieves 52.73% accuracy (+16.37%) and demonstrates performance comparable to GPT-4o on challenging tasks. To summarize, by enabling scalable generalization and modular domain transfer, our work establishes a foundation for the next generation of general-purpose AI assistants.
Robust Cross-View Geo-Localization via Content-Viewpoint Disentanglement
May 17, 2025Abstract:Cross-view geo-localization (CVGL) aims to match images of the same geographic location captured from different perspectives, such as drones and satellites. Despite recent advances, CVGL remains highly challenging due to significant appearance changes and spatial distortions caused by viewpoint variations. Existing methods typically assume that cross-view images can be directly aligned within a shared feature space by maximizing feature similarity through contrastive learning. Nonetheless, this assumption overlooks the inherent conflicts induced by viewpoint discrepancies, resulting in extracted features containing inconsistent information that hinders precise localization. In this study, we take a manifold learning perspective and model the feature space of cross-view images as a composite manifold jointly governed by content and viewpoint information. Building upon this insight, we propose $\textbf{CVD}$, a new CVGL framework that explicitly disentangles $\textit{content}$ and $\textit{viewpoint}$ factors. To promote effective disentanglement, we introduce two constraints: $\textit{(i)}$ An intra-view independence constraint, which encourages statistical independence between the two factors by minimizing their mutual information. $\textit{(ii)}$ An inter-view reconstruction constraint that reconstructs each view by cross-combining $\textit{content}$ and $\textit{viewpoint}$ from paired images, ensuring factor-specific semantics are preserved. As a plug-and-play module, CVD can be seamlessly integrated into existing geo-localization pipelines. Extensive experiments on four benchmarks, i.e., University-1652, SUES-200, CVUSA, and CVACT, demonstrate that CVD consistently improves both localization accuracy and generalization across multiple baselines.
SonicSieve: Bringing Directional Speech Extraction to Smartphones Using Acoustic Microstructures
Apr 15, 2025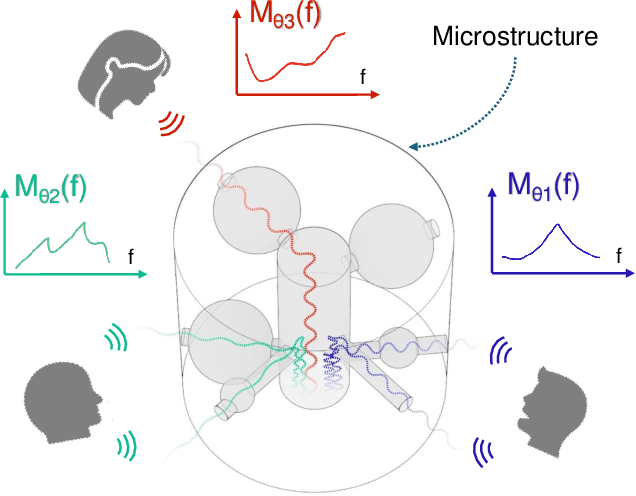
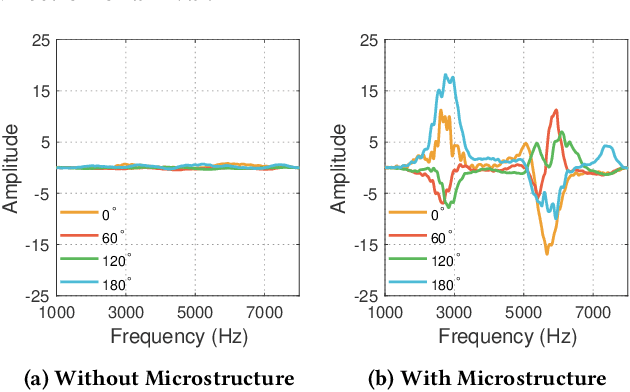
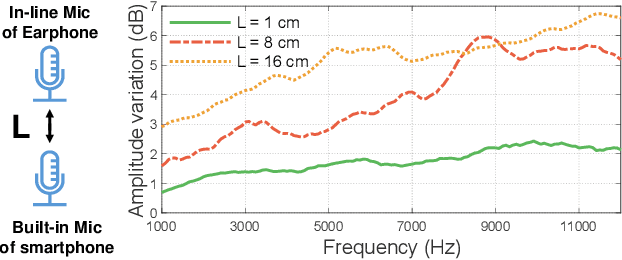
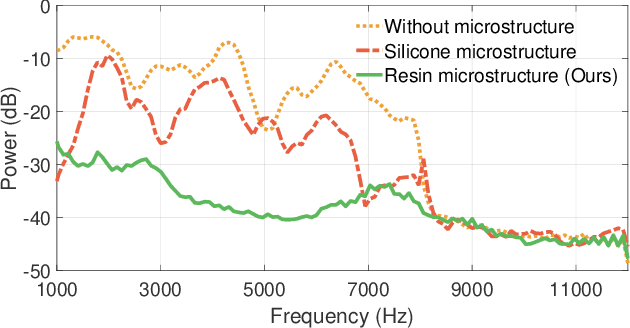
Abstract:Imagine placing your smartphone on a table in a noisy restaurant and clearly capturing the voices of friends seated around you, or recording a lecturer's voice with clarity in a reverberant auditorium. We introduce SonicSieve, the first intelligent directional speech extraction system for smartphones using a bio-inspired acoustic microstructure. Our passive design embeds directional cues onto incoming speech without any additional electronics. It attaches to the in-line mic of low-cost wired earphones which can be attached to smartphones. We present an end-to-end neural network that processes the raw audio mixtures in real-time on mobile devices. Our results show that SonicSieve achieves a signal quality improvement of 5.0 dB when focusing on a 30{\deg} angular region. Additionally, the performance of our system based on only two microphones exceeds that of conventional 5-microphone arrays.
Category Prompt Mamba Network for Nuclei Segmentation and Classification
Mar 13, 2025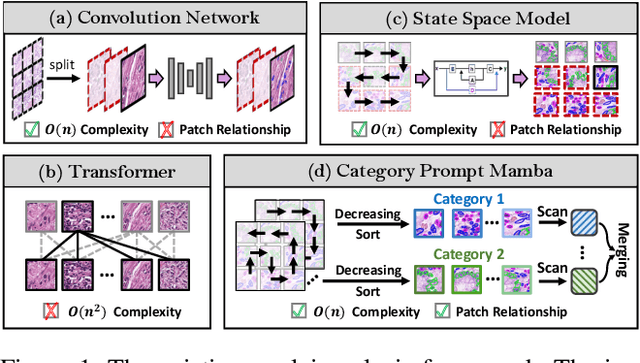
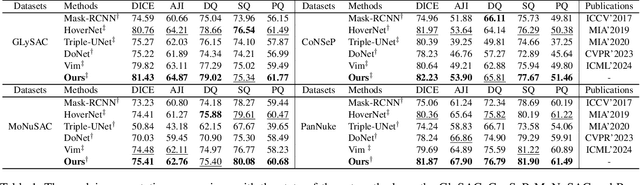
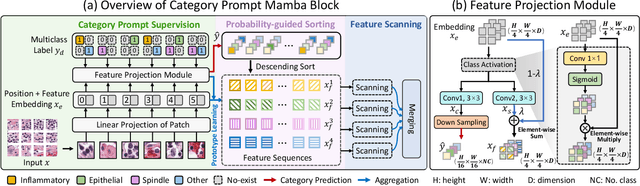
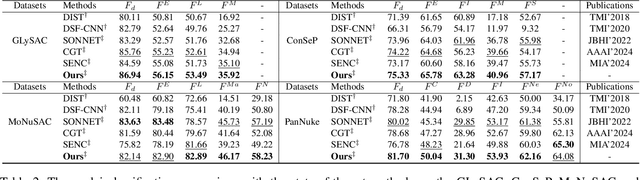
Abstract:Nuclei segmentation and classification provide an essential basis for tumor immune microenvironment analysis. The previous nuclei segmentation and classification models require splitting large images into smaller patches for training, leading to two significant issues. First, nuclei at the borders of adjacent patches often misalign during inference. Second, this patch-based approach significantly increases the model's training and inference time. Recently, Mamba has garnered attention for its ability to model large-scale images with linear time complexity and low memory consumption. It offers a promising solution for training nuclei segmentation and classification models on full-sized images. However, the Mamba orientation-based scanning method lacks account for category-specific features, resulting in sub-optimal performance in scenarios with imbalanced class distributions. To address these challenges, this paper introduces a novel scanning strategy based on category probability sorting, which independently ranks and scans features for each category according to confidence from high to low. This approach enhances the feature representation of uncertain samples and mitigates the issues caused by imbalanced distributions. Extensive experiments conducted on four public datasets demonstrate that our method outperforms state-of-the-art approaches, delivering superior performance in nuclei segmentation and classification tasks.
HEROS-GAN: Honed-Energy Regularized and Optimal Supervised GAN for Enhancing Accuracy and Range of Low-Cost Accelerometers
Feb 25, 2025Abstract:Low-cost accelerometers play a crucial role in modern society due to their advantages of small size, ease of integration, wearability, and mass production, making them widely applicable in automotive systems, aerospace, and wearable technology. However, this widely used sensor suffers from severe accuracy and range limitations. To this end, we propose a honed-energy regularized and optimal supervised GAN (HEROS-GAN), which transforms low-cost sensor signals into high-cost equivalents, thereby overcoming the precision and range limitations of low-cost accelerometers. Due to the lack of frame-level paired low-cost and high-cost signals for training, we propose an Optimal Transport Supervision (OTS), which leverages optimal transport theory to explore potential consistency between unpaired data, thereby maximizing supervisory information. Moreover, we propose a Modulated Laplace Energy (MLE), which injects appropriate energy into the generator to encourage it to break range limitations, enhance local changes, and enrich signal details. Given the absence of a dedicated dataset, we specifically establish a Low-cost Accelerometer Signal Enhancement Dataset (LASED) containing tens of thousands of samples, which is the first dataset serving to improve the accuracy and range of accelerometers and is released in Github. Experimental results demonstrate that a GAN combined with either OTS or MLE alone can surpass the previous signal enhancement SOTA methods by an order of magnitude. Integrating both OTS and MLE, the HEROS-GAN achieves remarkable results, which doubles the accelerometer range while reducing signal noise by two orders of magnitude, establishing a benchmark in the accelerometer signal processing.
HisynSeg: Weakly-Supervised Histopathological Image Segmentation via Image-Mixing Synthesis and Consistency Regularization
Dec 30, 2024



Abstract:Tissue semantic segmentation is one of the key tasks in computational pathology. To avoid the expensive and laborious acquisition of pixel-level annotations, a wide range of studies attempt to adopt the class activation map (CAM), a weakly-supervised learning scheme, to achieve pixel-level tissue segmentation. However, CAM-based methods are prone to suffer from under-activation and over-activation issues, leading to poor segmentation performance. To address this problem, we propose a novel weakly-supervised semantic segmentation framework for histopathological images based on image-mixing synthesis and consistency regularization, dubbed HisynSeg. Specifically, synthesized histopathological images with pixel-level masks are generated for fully-supervised model training, where two synthesis strategies are proposed based on Mosaic transformation and B\'ezier mask generation. Besides, an image filtering module is developed to guarantee the authenticity of the synthesized images. In order to further avoid the model overfitting to the occasional synthesis artifacts, we additionally propose a novel self-supervised consistency regularization, which enables the real images without segmentation masks to supervise the training of the segmentation model. By integrating the proposed techniques, the HisynSeg framework successfully transforms the weakly-supervised semantic segmentation problem into a fully-supervised one, greatly improving the segmentation accuracy. Experimental results on three datasets prove that the proposed method achieves a state-of-the-art performance. Code is available at https://github.com/Vison307/HisynSeg.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge