Category Prompt Mamba Network for Nuclei Segmentation and Classification
Paper and Code
Mar 13, 2025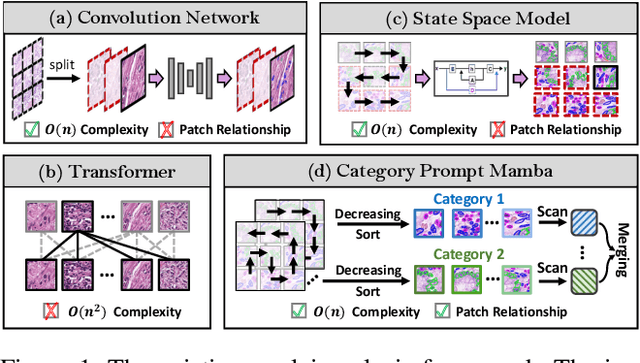
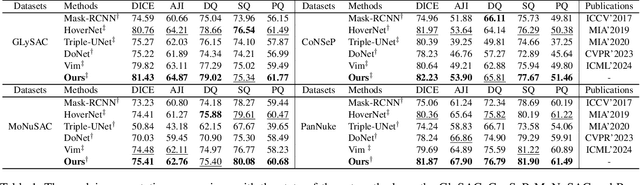
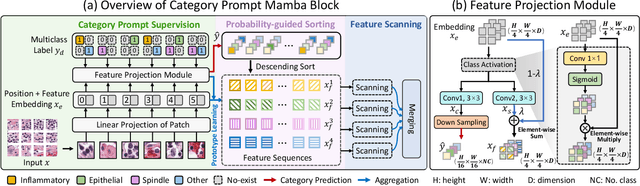
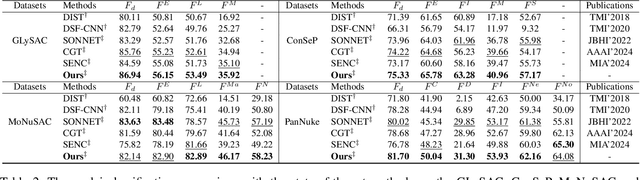
Nuclei segmentation and classification provide an essential basis for tumor immune microenvironment analysis. The previous nuclei segmentation and classification models require splitting large images into smaller patches for training, leading to two significant issues. First, nuclei at the borders of adjacent patches often misalign during inference. Second, this patch-based approach significantly increases the model's training and inference time. Recently, Mamba has garnered attention for its ability to model large-scale images with linear time complexity and low memory consumption. It offers a promising solution for training nuclei segmentation and classification models on full-sized images. However, the Mamba orientation-based scanning method lacks account for category-specific features, resulting in sub-optimal performance in scenarios with imbalanced class distributions. To address these challenges, this paper introduces a novel scanning strategy based on category probability sorting, which independently ranks and scans features for each category according to confidence from high to low. This approach enhances the feature representation of uncertain samples and mitigates the issues caused by imbalanced distributions. Extensive experiments conducted on four public datasets demonstrate that our method outperforms state-of-the-art approaches, delivering superior performance in nuclei segmentation and classification tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge