Jianxu Chen
Toward Auditable Neuro-Symbolic Reasoning in Pathology: SQL as an Explicit Trace of Evidence
Jan 05, 2026Abstract:Automated pathology image analysis is central to clinical diagnosis, but clinicians still ask which slide features drive a model's decision and why. Vision-language models can produce natural language explanations, but these are often correlational and lack verifiable evidence. In this paper, we introduce an SQL-centered agentic framework that enables both feature measurement and reasoning to be auditable. Specifically, after extracting human-interpretable cellular features, Feature Reasoning Agents compose and execute SQL queries over feature tables to aggregate visual evidence into quantitative findings. A Knowledge Comparison Agent then evaluates these findings against established pathological knowledge, mirroring how pathologists justify diagnoses from measurable observations. Extensive experiments evaluated on two pathology visual question answering datasets demonstrate our method improves interpretability and decision traceability while producing executable SQL traces that link cellular measurements to diagnostic conclusions.
FlowRoI A Fast Optical Flow Driven Region of Interest Extraction Framework for High-Throughput Image Compression in Immune Cell Migration Analysis
Nov 18, 2025Abstract:Autonomous migration is essential for the function of immune cells such as neutrophils and plays a pivotal role in diverse diseases. Recently, we introduced ComplexEye, a multi-lens array microscope comprising 16 independent aberration-corrected glass lenses arranged at the pitch of a 96-well plate, capable of capturing high-resolution movies of migrating cells. This architecture enables high-throughput live-cell video microscopy for migration analysis, supporting routine quantification of autonomous motility with strong potential for clinical translation. However, ComplexEye and similar high-throughput imaging platforms generate data at an exponential rate, imposing substantial burdens on storage and transmission. To address this challenge, we present FlowRoI, a fast optical-flow-based region of interest (RoI) extraction framework designed for high-throughput image compression in immune cell migration studies. FlowRoI estimates optical flow between consecutive frames and derives RoI masks that reliably cover nearly all migrating cells. The raw image and its corresponding RoI mask are then jointly encoded using JPEG2000 to enable RoI-aware compression. FlowRoI operates with high computational efficiency, achieving runtimes comparable to standard JPEG2000 and reaching an average throughput of about 30 frames per second on a modern laptop equipped with an Intel i7-1255U CPU. In terms of image quality, FlowRoI yields higher peak signal-to-noise ratio (PSNR) in cellular regions and achieves 2.0-2.2x higher compression rates at matched PSNR compared to standard JPEG2000.
An Active Learning Pipeline for Biomedical Image Instance Segmentation with Minimal Human Intervention
Nov 06, 2025Abstract:Biomedical image segmentation is critical for precise structure delineation and downstream analysis. Traditional methods often struggle with noisy data, while deep learning models such as U-Net have set new benchmarks in segmentation performance. nnU-Net further automates model configuration, making it adaptable across datasets without extensive tuning. However, it requires a substantial amount of annotated data for cross-validation, posing a challenge when only raw images but no labels are available. Large foundation models offer zero-shot generalizability, but may underperform on specific datasets with unique characteristics, limiting their direct use for analysis. This work addresses these bottlenecks by proposing a data-centric AI workflow that leverages active learning and pseudo-labeling to combine the strengths of traditional neural networks and large foundation models while minimizing human intervention. The pipeline starts by generating pseudo-labels from a foundation model, which are then used for nnU-Net's self-configuration. Subsequently, a representative core-set is selected for minimal manual annotation, enabling effective fine-tuning of the nnU-Net model. This approach significantly reduces the need for manual annotations while maintaining competitive performance, providing an accessible solution for biomedical researchers to apply state-of-the-art AI techniques in their segmentation tasks. The code is available at https://github.com/MMV-Lab/AL_BioMed_img_seg.
* 6 pages, 4 figures, presented at Bildverarbeitung f\"ur die Medizin (BVM) 2025, Wiesbaden, Germany
Data Efficiency and Transfer Robustness in Biomedical Image Segmentation: A Study of Redundancy and Forgetting with Cellpose
Nov 06, 2025Abstract:Generalist biomedical image segmentation models such as Cellpose are increasingly applied across diverse imaging modalities and cell types. However, two critical challenges remain underexplored: (1) the extent of training data redundancy and (2) the impact of cross domain transfer on model retention. In this study, we conduct a systematic empirical analysis of these challenges using Cellpose as a case study. First, to assess data redundancy, we propose a simple dataset quantization (DQ) strategy for constructing compact yet diverse training subsets. Experiments on the Cyto dataset show that image segmentation performance saturates with only 10% of the data, revealing substantial redundancy and potential for training with minimal annotations. Latent space analysis using MAE embeddings and t-SNE confirms that DQ selected patches capture greater feature diversity than random sampling. Second, to examine catastrophic forgetting, we perform cross domain finetuning experiments and observe significant degradation in source domain performance, particularly when adapting from generalist to specialist domains. We demonstrate that selective DQ based replay reintroducing just 5-10% of the source data effectively restores source performance, while full replay can hinder target adaptation. Additionally, we find that training domain sequencing improves generalization and reduces forgetting in multi stage transfer. Our findings highlight the importance of data centric design in biomedical image segmentation and suggest that efficient training requires not only compact subsets but also retention aware learning strategies and informed domain ordering. The code is available at https://github.com/MMV-Lab/biomedseg-efficiency.
PathMR: Multimodal Visual Reasoning for Interpretable Pathology Diagnosis
Aug 28, 2025Abstract:Deep learning based automated pathological diagnosis has markedly improved diagnostic efficiency and reduced variability between observers, yet its clinical adoption remains limited by opaque model decisions and a lack of traceable rationale. To address this, recent multimodal visual reasoning architectures provide a unified framework that generates segmentation masks at the pixel level alongside semantically aligned textual explanations. By localizing lesion regions and producing expert style diagnostic narratives, these models deliver the transparent and interpretable insights necessary for dependable AI assisted pathology. Building on these advancements, we propose PathMR, a cell-level Multimodal visual Reasoning framework for Pathological image analysis. Given a pathological image and a textual query, PathMR generates expert-level diagnostic explanations while simultaneously predicting cell distribution patterns. To benchmark its performance, we evaluated our approach on the publicly available PathGen dataset as well as on our newly developed GADVR dataset. Extensive experiments on these two datasets demonstrate that PathMR consistently outperforms state-of-the-art visual reasoning methods in text generation quality, segmentation accuracy, and cross-modal alignment. These results highlight the potential of PathMR for improving interpretability in AI-driven pathological diagnosis. The code will be publicly available in https://github.com/zhangye-zoe/PathMR.
The Four Color Theorem for Cell Instance Segmentation
Jun 11, 2025Abstract:Cell instance segmentation is critical to analyzing biomedical images, yet accurately distinguishing tightly touching cells remains a persistent challenge. Existing instance segmentation frameworks, including detection-based, contour-based, and distance mapping-based approaches, have made significant progress, but balancing model performance with computational efficiency remains an open problem. In this paper, we propose a novel cell instance segmentation method inspired by the four-color theorem. By conceptualizing cells as countries and tissues as oceans, we introduce a four-color encoding scheme that ensures adjacent instances receive distinct labels. This reformulation transforms instance segmentation into a constrained semantic segmentation problem with only four predicted classes, substantially simplifying the instance differentiation process. To solve the training instability caused by the non-uniqueness of four-color encoding, we design an asymptotic training strategy and encoding transformation method. Extensive experiments on various modes demonstrate our approach achieves state-of-the-art performance. The code is available at https://github.com/zhangye-zoe/FCIS.
Adaptive Kinematic Modeling for Improved Hand Posture Estimates Using a Haptic Glove
Nov 10, 2024Abstract:Most commercially available haptic gloves compromise the accuracy of hand-posture measurements in favor of a simpler design with fewer sensors. While inaccurate posture data is often sufficient for the task at hand in biomedical settings such as VR-therapy-aided rehabilitation, measurements should be as precise as possible to digitally recreate hand postures as accurately as possible. With these applications in mind, we have added extra sensors to the commercially available Dexmo haptic glove by Dexta Robotics and applied kinematic models of the haptic glove and the user's hand to improve the accuracy of hand-posture measurements. In this work, we describe the augmentations and the kinematic modeling approach. Additionally, we present and discuss an evaluation of hand posture measurements as a proof of concept.
Rethinking Medical Anomaly Detection in Brain MRI: An Image Quality Assessment Perspective
Aug 15, 2024



Abstract:Reconstruction-based methods, particularly those leveraging autoencoders, have been widely adopted to perform anomaly detection in brain MRI. While most existing works try to improve detection accuracy by proposing new model structures or algorithms, we tackle the problem through image quality assessment, an underexplored perspective in the field. We propose a fusion quality loss function that combines Structural Similarity Index Measure loss with l1 loss, offering a more comprehensive evaluation of reconstruction quality. Additionally, we introduce a data pre-processing strategy that enhances the average intensity ratio (AIR) between normal and abnormal regions, further improving the distinction of anomalies. By fusing the aforementioned two methods, we devise the image quality assessment (IQA) approach. The proposed IQA approach achieves significant improvements (>10%) in terms of Dice coefficient (DICE) and Area Under the Precision-Recall Curve (AUPRC) on the BraTS21 (T2, FLAIR) and MSULB datasets when compared with state-of-the-art methods. These results highlight the importance of invoking the comprehensive image quality assessment in medical anomaly detection and provide a new perspective for future research in this field.
Multimodal Large Language Models for Bioimage Analysis
Jul 29, 2024Abstract:Rapid advancements in imaging techniques and analytical methods over the past decade have revolutionized our ability to comprehensively probe the biological world at multiple scales, pinpointing the type, quantity, location, and even temporal dynamics of biomolecules. The surge in data complexity and volume presents significant challenges in translating this wealth of information into knowledge. The recently emerged Multimodal Large Language Models (MLLMs) exhibit strong emergent capacities, such as understanding, analyzing, reasoning, and generalization. With these capabilities, MLLMs hold promise to extract intricate information from biological images and data obtained through various modalities, thereby expediting our biological understanding and aiding in the development of novel computational frameworks. Previously, such capabilities were mostly attributed to humans for interpreting and summarizing meaningful conclusions from comprehensive observations and analysis of biological images. However, the current development of MLLMs shows increasing promise in serving as intelligent assistants or agents for augmenting human researchers in biology research
Implicit Neural Image Field for Biological Microscopy Image Compression
May 29, 2024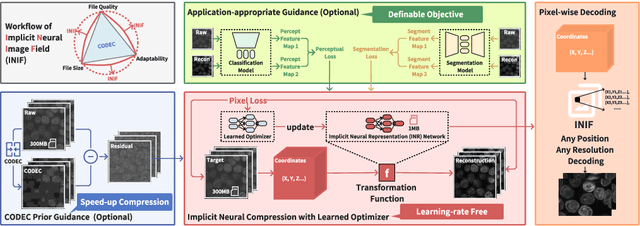
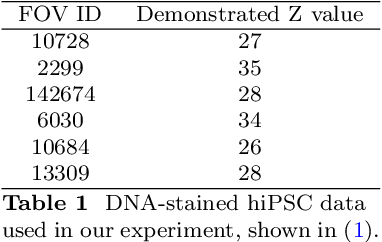
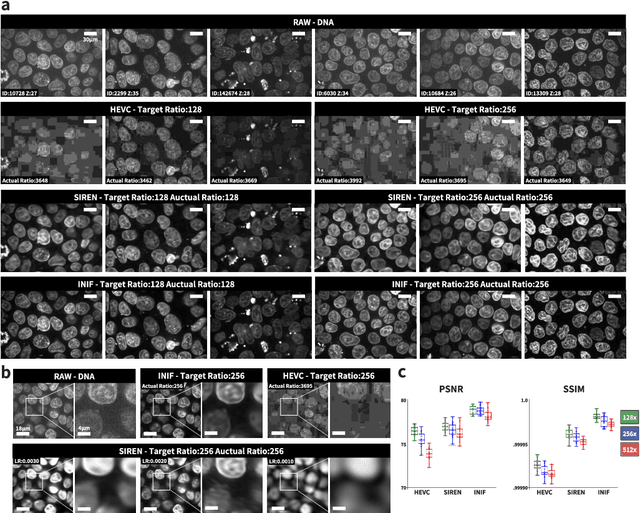
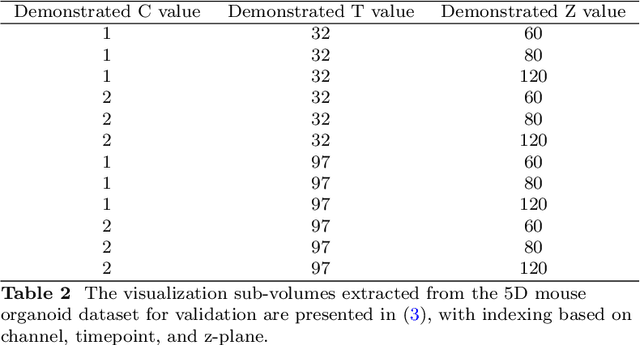
Abstract:The rapid pace of innovation in biological microscopy imaging has led to large images, putting pressure on data storage and impeding efficient sharing, management, and visualization. This necessitates the development of efficient compression solutions. Traditional CODEC methods struggle to adapt to the diverse bioimaging data and often suffer from sub-optimal compression. In this study, we propose an adaptive compression workflow based on Implicit Neural Representation (INR). This approach permits application-specific compression objectives, capable of compressing images of any shape and arbitrary pixel-wise decompression. We demonstrated on a wide range of microscopy images from real applications that our workflow not only achieved high, controllable compression ratios (e.g., 512x) but also preserved detailed information critical for downstream analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge