Antonio Pepe
Efficient MedSAMs: Segment Anything in Medical Images on Laptop
Dec 20, 2024


Abstract:Promptable segmentation foundation models have emerged as a transformative approach to addressing the diverse needs in medical images, but most existing models require expensive computing, posing a big barrier to their adoption in clinical practice. In this work, we organized the first international competition dedicated to promptable medical image segmentation, featuring a large-scale dataset spanning nine common imaging modalities from over 20 different institutions. The top teams developed lightweight segmentation foundation models and implemented an efficient inference pipeline that substantially reduced computational requirements while maintaining state-of-the-art segmentation accuracy. Moreover, the post-challenge phase advanced the algorithms through the design of performance booster and reproducibility tasks, resulting in improved algorithms and validated reproducibility of the winning solution. Furthermore, the best-performing algorithms have been incorporated into the open-source software with a user-friendly interface to facilitate clinical adoption. The data and code are publicly available to foster the further development of medical image segmentation foundation models and pave the way for impactful real-world applications.
Deep Medial Voxels: Learned Medial Axis Approximations for Anatomical Shape Modeling
Mar 18, 2024Abstract:Shape reconstruction from imaging volumes is a recurring need in medical image analysis. Common workflows start with a segmentation step, followed by careful post-processing and,finally, ad hoc meshing algorithms. As this sequence can be timeconsuming, neural networks are trained to reconstruct shapes through template deformation. These networks deliver state-ofthe-art results without manual intervention, but, so far, they have primarily been evaluated on anatomical shapes with little topological variety between individuals. In contrast, other works favor learning implicit shape models, which have multiple benefits for meshing and visualization. Our work follows this direction by introducing deep medial voxels, a semi-implicit representation that faithfully approximates the topological skeleton from imaging volumes and eventually leads to shape reconstruction via convolution surfaces. Our reconstruction technique shows potential for both visualization and computer simulations.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
Anatomy Completor: A Multi-class Completion Framework for 3D Anatomy Reconstruction
Sep 10, 2023Abstract:In this paper, we introduce a completion framework to reconstruct the geometric shapes of various anatomies, including organs, vessels and muscles. Our work targets a scenario where one or multiple anatomies are missing in the imaging data due to surgical, pathological or traumatic factors, or simply because these anatomies are not covered by image acquisition. Automatic reconstruction of the missing anatomies benefits many applications, such as organ 3D bio-printing, whole-body segmentation, animation realism, paleoradiology and forensic imaging. We propose two paradigms based on a 3D denoising auto-encoder (DAE) to solve the anatomy reconstruction problem: (i) the DAE learns a many-to-one mapping between incomplete and complete instances; (ii) the DAE learns directly a one-to-one residual mapping between the incomplete instances and the target anatomies. We apply a loss aggregation scheme that enables the DAE to learn the many-to-one mapping more effectively and further enhances the learning of the residual mapping. On top of this, we extend the DAE to a multiclass completor by assigning a unique label to each anatomy involved. We evaluate our method using a CT dataset with whole-body segmentations. Results show that our method produces reasonable anatomy reconstructions given instances with different levels of incompleteness (i.e., one or multiple random anatomies are missing). Codes and pretrained models are publicly available at https://github.com/Jianningli/medshapenet-feedback/ tree/main/anatomy-completor
The HoloLens in Medicine: A systematic Review and Taxonomy
Sep 06, 2022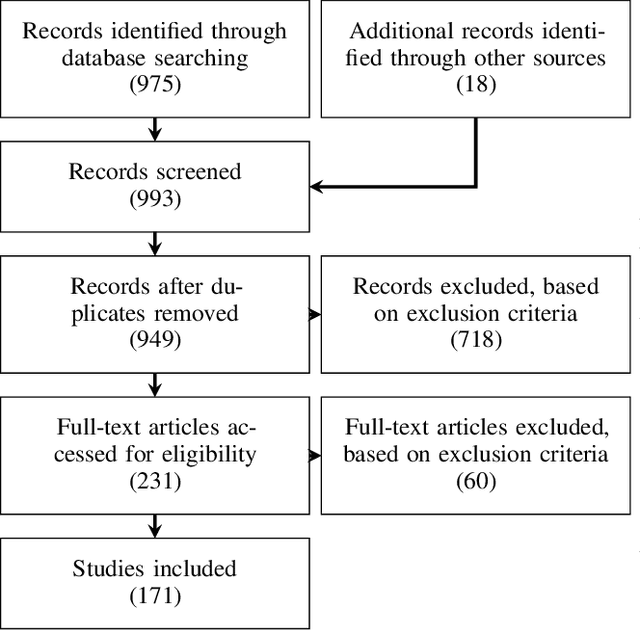
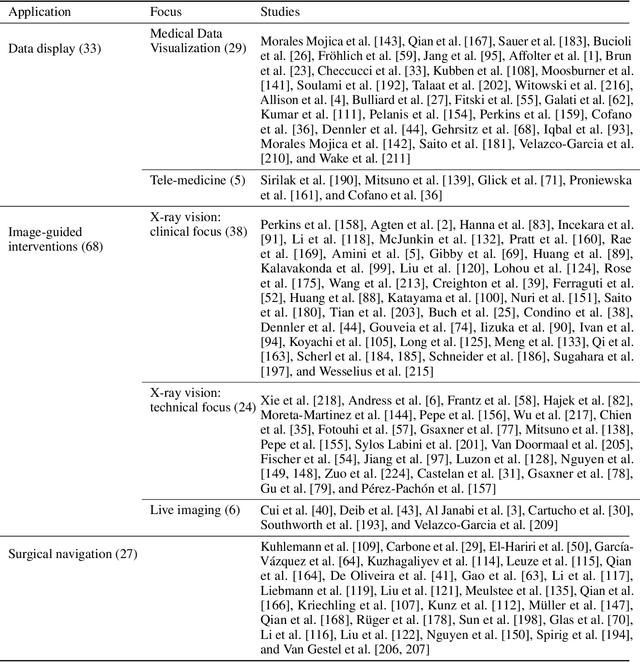
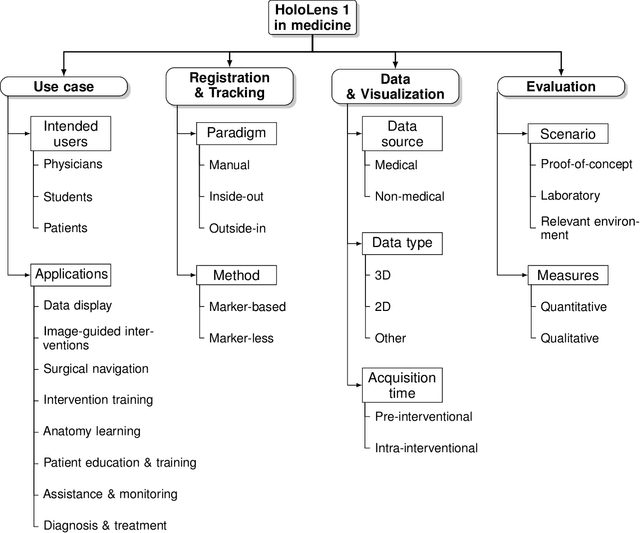

Abstract:The HoloLens (Microsoft Corp., Redmond, WA), a head-worn, optically see-through augmented reality display, is the main player in the recent boost in medical augmented reality research. In medical settings, the HoloLens enables the physician to obtain immediate insight into patient information, directly overlaid with their view of the clinical scenario, the medical student to gain a better understanding of complex anatomies or procedures, and even the patient to execute therapeutic tasks with improved, immersive guidance. In this systematic review, we provide a comprehensive overview of the usage of the first-generation HoloLens within the medical domain, from its release in March 2016, until the year of 2021, were attention is shifting towards it's successor, the HoloLens 2. We identified 171 relevant publications through a systematic search of the PubMed and Scopus databases. We analyze these publications in regard to their intended use case, technical methodology for registration and tracking, data sources, visualization as well as validation and evaluation. We find that, although the feasibility of using the HoloLens in various medical scenarios has been shown, increased efforts in the areas of precision, reliability, usability, workflow and perception are necessary to establish AR in clinical practice.
Back to the Roots: Reconstructing Large and Complex Cranial Defects using an Image-based Statistical Shape Model
Apr 12, 2022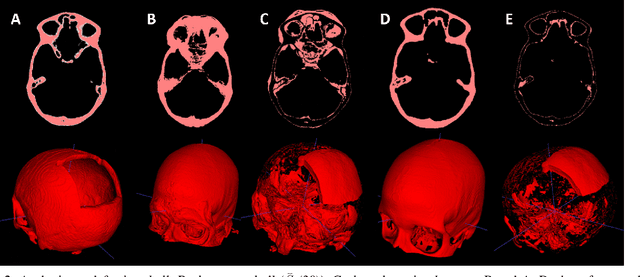
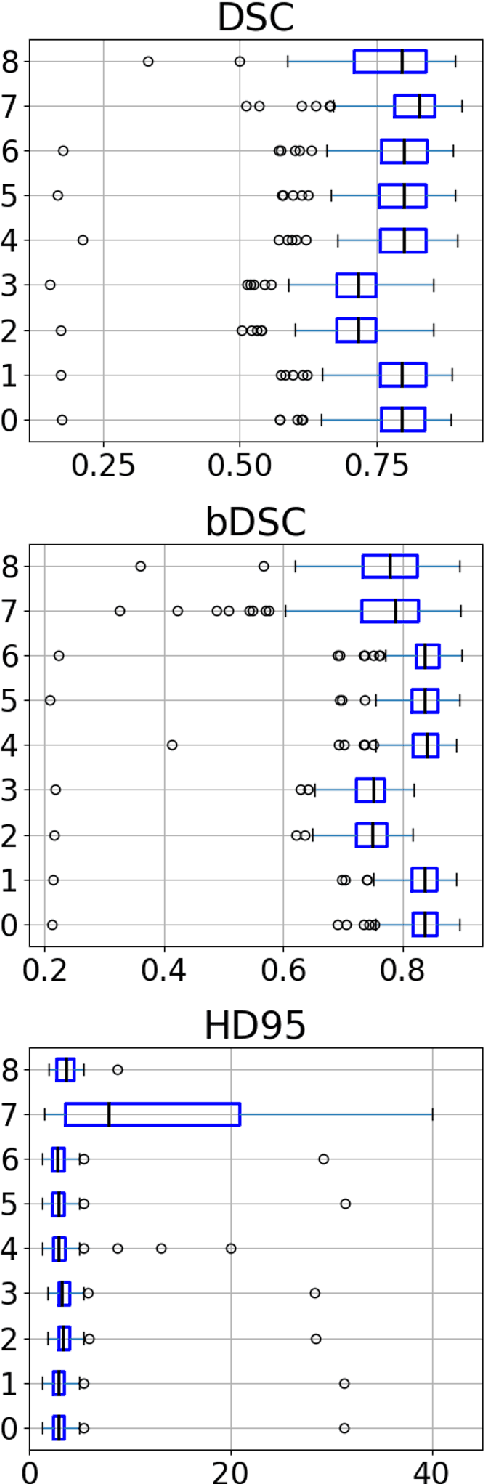
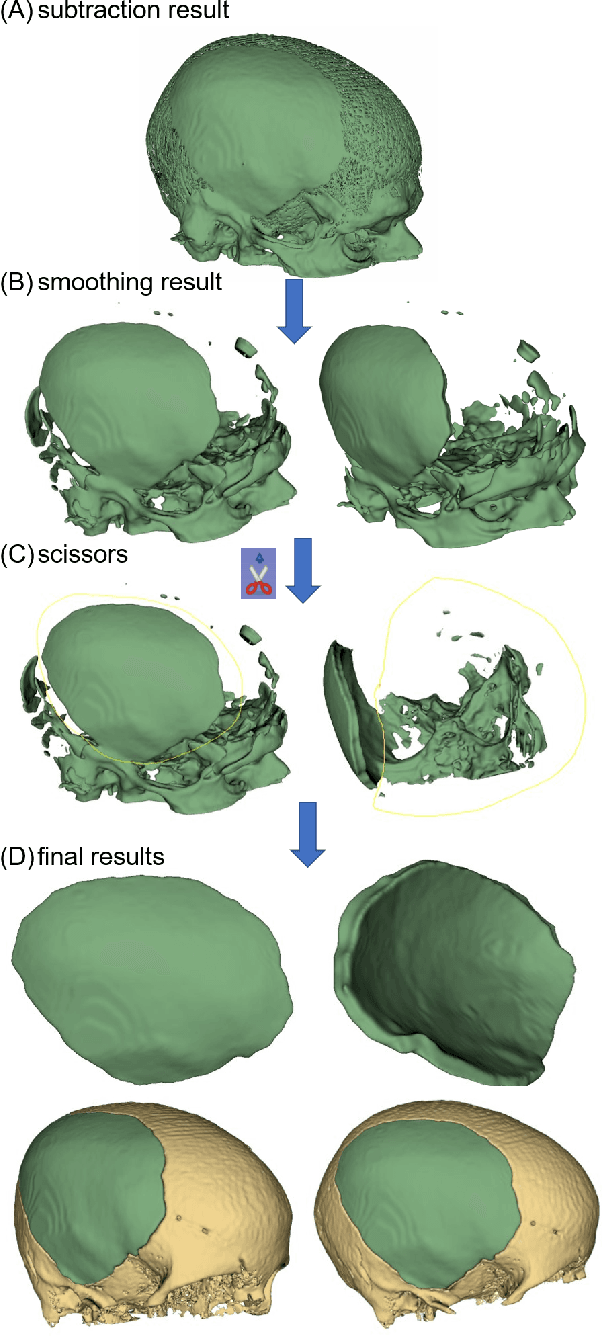
Abstract:Designing implants for large and complex cranial defects is a challenging task, even for professional designers. Current efforts on automating the design process focused mainly on convolutional neural networks (CNN), which have produced state-of-the-art results on reconstructing synthetic defects. However, existing CNN-based methods have been difficult to translate to clinical practice in cranioplasty, as their performance on complex and irregular cranial defects remains unsatisfactory. In this paper, a statistical shape model (SSM) built directly on the segmentation masks of the skulls is presented. We evaluate the SSM on several cranial implant design tasks, and the results show that, while the SSM performs suboptimally on synthetic defects compared to CNN-based approaches, it is capable of reconstructing large and complex defects with only minor manual corrections. The quality of the resulting implants is examined and assured by experienced neurosurgeons. In contrast, CNN-based approaches, even with massive data augmentation, fail or produce less-than-satisfactory implants for these cases. Codes are publicly available at https://github.com/Jianningli/ssm
Stochastic Modeling of Inhomogeneities in the Aortic Wall and Uncertainty Quantification using a Bayesian Encoder-Decoder Surrogate
Feb 21, 2022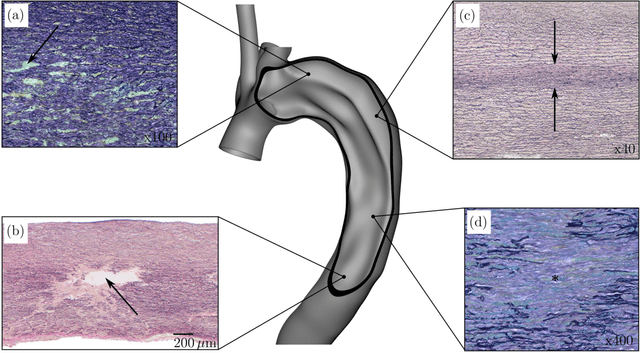
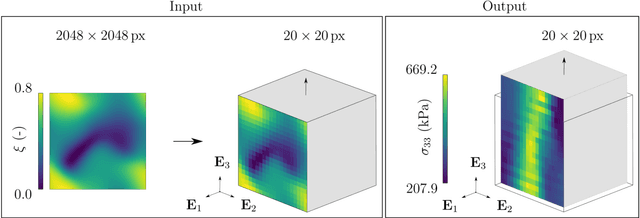
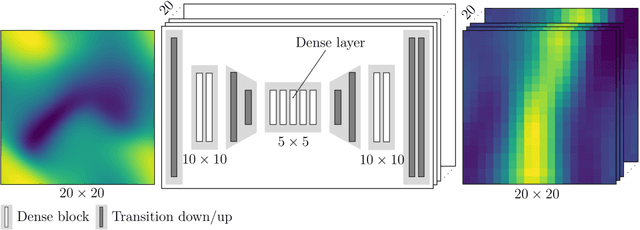
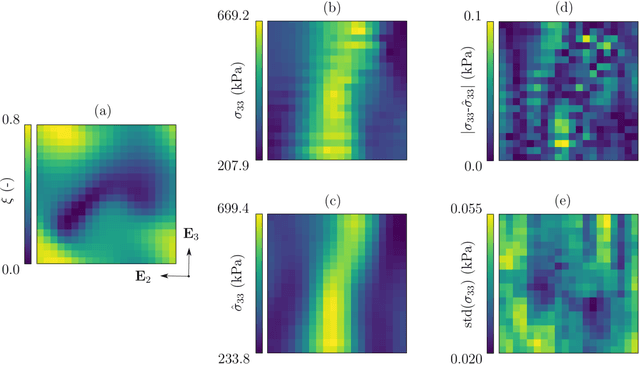
Abstract:Inhomogeneities in the aortic wall can lead to localized stress accumulations, possibly initiating dissection. In many cases, a dissection results from pathological changes such as fragmentation or loss of elastic fibers. But it has been shown that even the healthy aortic wall has an inherent heterogeneous microstructure. Some parts of the aorta are particularly susceptible to the development of inhomogeneities due to pathological changes, however, the distribution in the aortic wall and the spatial extent, such as size, shape, and type, are difficult to predict. Motivated by this observation, we describe the heterogeneous distribution of elastic fiber degradation in the dissected aortic wall using a stochastic constitutive model. For this purpose, random field realizations, which model the stochastic distribution of degraded elastic fibers, are generated over a non-equidistant grid. The random field then serves as input for a uni-axial extension test of the pathological aortic wall, solved with the finite-element (FE) method. To include the microstructure of the dissected aortic wall, a constitutive model developed in a previous study is applied, which also includes an approach to model the degradation of inter-lamellar elastic fibers. Then to assess the uncertainty in the output stress distribution due to this stochastic constitutive model, a convolutional neural network, specifically a Bayesian encoder-decoder, was used as a surrogate model that maps the random input fields to the output stress distribution obtained from the FE analysis. The results show that the neural network is able to predict the stress distribution of the FE analysis while significantly reducing the computational time. In addition, it provides the probability for exceeding critical stresses within the aortic wall, which could allow for the prediction of delamination or fatal rupture.
Automated cross-sectional view selection in CT angiography of aortic dissections with uncertainty awareness and retrospective clinical annotations
Nov 22, 2021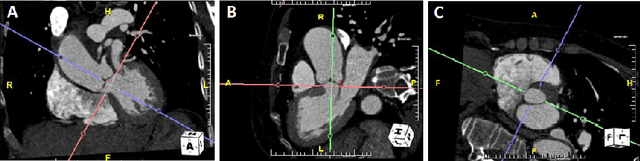
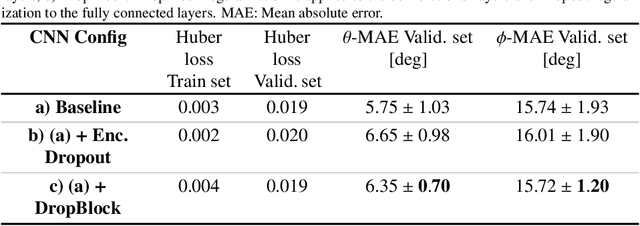
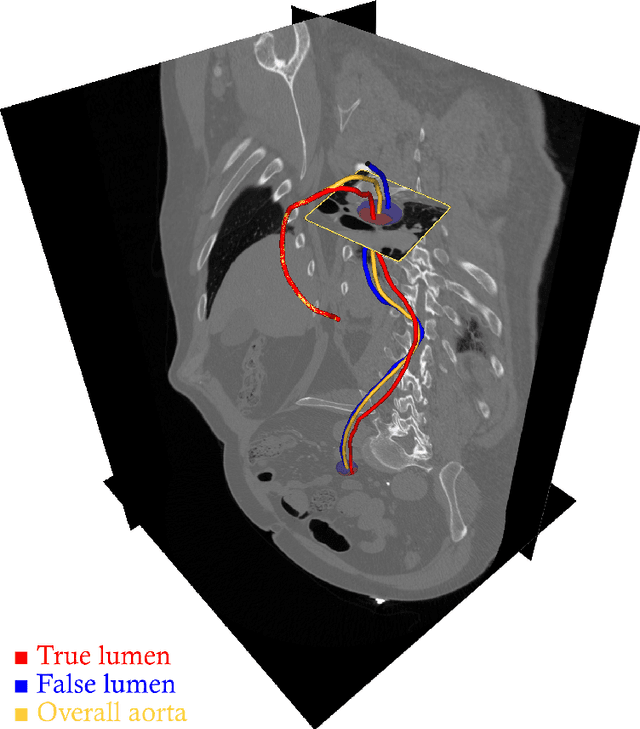
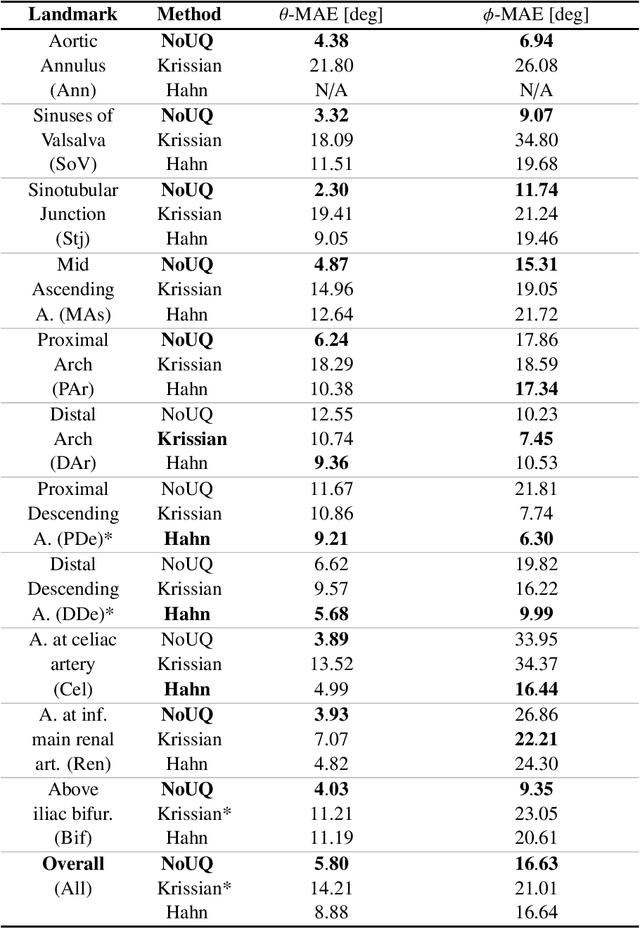
Abstract:Objective: Surveillance imaging of chronic aortic diseases, such as dissections, relies on obtaining and comparing cross-sectional diameter measurements at predefined aortic landmarks, over time. Due to a lack of robust tools, the orientation of the cross-sectional planes is defined manually by highly trained operators. We show how manual annotations routinely collected in a clinic can be efficiently used to ease this task, despite the presence of a non-negligible interoperator variability in the measurements. Impact: Ill-posed but repetitive imaging tasks can be eased or automated by leveraging imperfect, retrospective clinical annotations. Methodology: In this work, we combine convolutional neural networks and uncertainty quantification methods to predict the orientation of such cross-sectional planes. We use clinical data randomly processed by 11 operators for training, and test on a smaller set processed by 3 independent operators to assess interoperator variability. Results: Our analysis shows that manual selection of cross-sectional planes is characterized by 95% limits of agreement (LOA) of $10.6^\circ$ and $21.4^\circ$ per angle. Our method showed to decrease static error by $3.57^\circ$ ($40.2$%) and $4.11^\circ$ ($32.8$%) against state of the art and LOA by $5.4^\circ$ ($49.0$%) and $16.0^\circ$ ($74.6$%) against manual processing. Conclusion: This suggests that pre-existing annotations can be an inexpensive resource in clinics to ease ill-posed and repetitive tasks like cross-section extraction for surveillance of aortic dissections.
Learning to Rearrange Voxels in Binary Segmentation Masks for Smooth Manifold Triangulation
Aug 11, 2021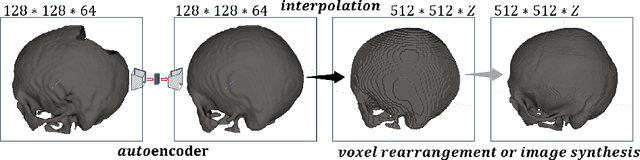
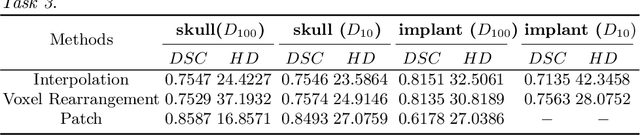
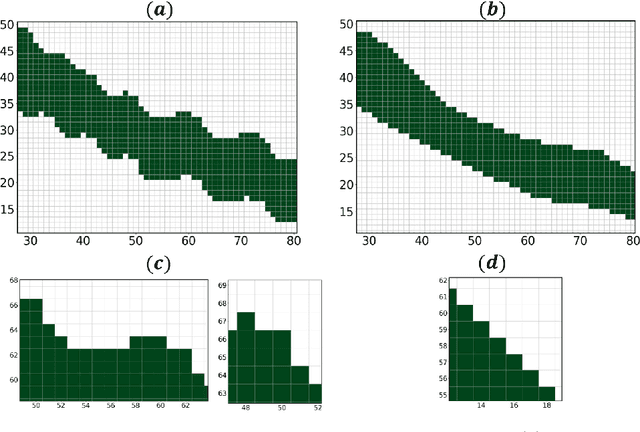
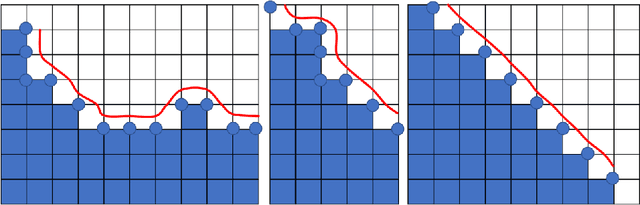
Abstract:Medical images, especially volumetric images, are of high resolution and often exceed the capacity of standard desktop GPUs. As a result, most deep learning-based medical image analysis tasks require the input images to be downsampled, often substantially, before these can be fed to a neural network. However, downsampling can lead to a loss of image quality, which is undesirable especially in reconstruction tasks, where the fine geometric details need to be preserved. In this paper, we propose that high-resolution images can be reconstructed in a coarse-to-fine fashion, where a deep learning algorithm is only responsible for generating a coarse representation of the image, which consumes moderate GPU memory. For producing the high-resolution outcome, we propose two novel methods: learned voxel rearrangement of the coarse output and hierarchical image synthesis. Compared to the coarse output, the high-resolution counterpart allows for smooth surface triangulation, which can be 3D-printed in the highest possible quality. Experiments of this paper are carried out on the dataset of AutoImplant 2021 (https://autoimplant2021.grand-challenge.org/), a MICCAI challenge on cranial implant design. The dataset contains high-resolution skulls that can be viewed as 2D manifolds embedded in a 3D space. Codes associated with this study can be accessed at https://github.com/Jianningli/voxel_rearrangement.
AI-based Aortic Vessel Tree Segmentation for Cardiovascular Diseases Treatment: Status Quo
Aug 06, 2021
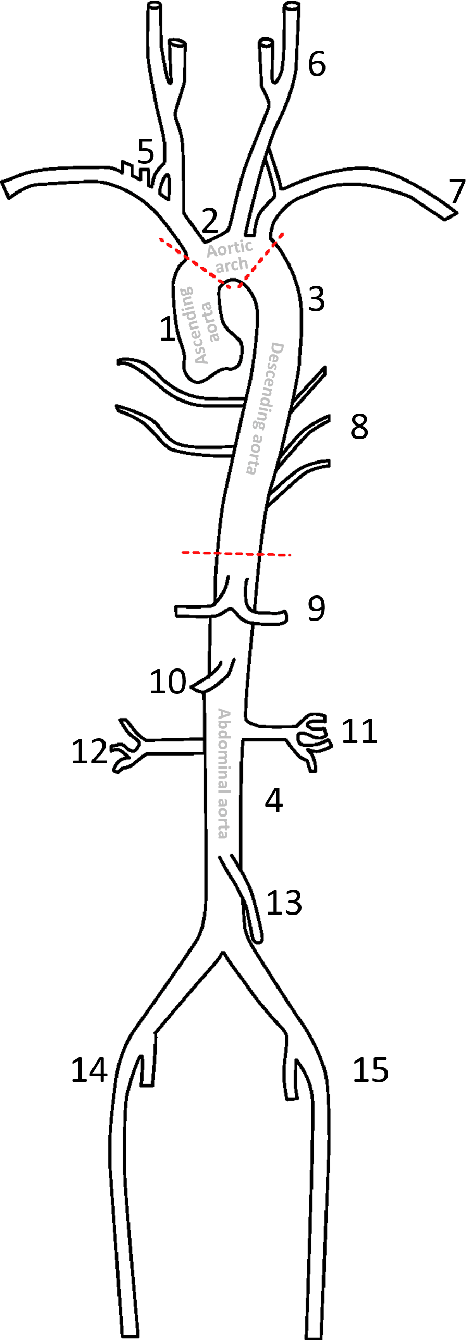
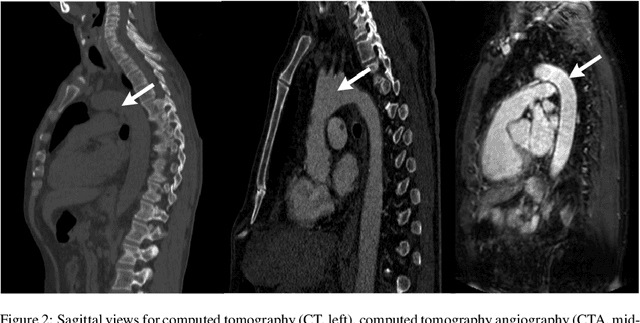
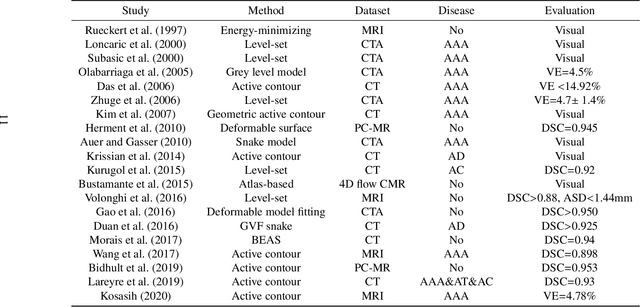
Abstract:The aortic vessel tree is composed of the aorta and its branching arteries, and plays a key role in supplying the whole body with blood. Aortic diseases, like aneurysms or dissections, can lead to an aortic rupture, whose treatment with open surgery is highly risky. Therefore, patients commonly undergo drug treatment under constant monitoring, which requires regular inspections of the vessels through imaging. The standard imaging modality for diagnosis and monitoring is computed tomography (CT), which can provide a detailed picture of the aorta and its branching vessels if combined with a contrast agent, resulting in a CT angiography (CTA). Optimally, the whole aortic vessel tree geometry from consecutive CTAs, are overlaid and compared. This allows to not only detect changes in the aorta, but also more peripheral vessel tree changes, caused by the primary pathology or newly developed. When performed manually, this reconstruction requires slice by slice contouring, which could easily take a whole day for a single aortic vessel tree and, hence, is not feasible in clinical practice. Automatic or semi-automatic vessel tree segmentation algorithms, on the other hand, can complete this task in a fraction of the manual execution time and run in parallel to the clinical routine of the clinicians. In this paper, we systematically review computing techniques for the automatic and semi-automatic segmentation of the aortic vessel tree. The review concludes with an in-depth discussion on how close these state-of-the-art approaches are to an application in clinical practice and how active this research field is, taking into account the number of publications, datasets and challenges.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge