Learning to Rearrange Voxels in Binary Segmentation Masks for Smooth Manifold Triangulation
Paper and Code
Aug 11, 2021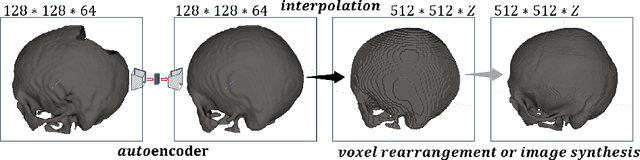
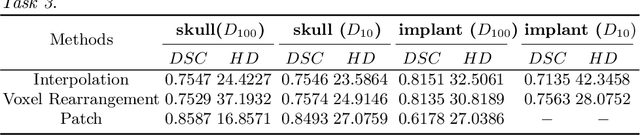
Medical images, especially volumetric images, are of high resolution and often exceed the capacity of standard desktop GPUs. As a result, most deep learning-based medical image analysis tasks require the input images to be downsampled, often substantially, before these can be fed to a neural network. However, downsampling can lead to a loss of image quality, which is undesirable especially in reconstruction tasks, where the fine geometric details need to be preserved. In this paper, we propose that high-resolution images can be reconstructed in a coarse-to-fine fashion, where a deep learning algorithm is only responsible for generating a coarse representation of the image, which consumes moderate GPU memory. For producing the high-resolution outcome, we propose two novel methods: learned voxel rearrangement of the coarse output and hierarchical image synthesis. Compared to the coarse output, the high-resolution counterpart allows for smooth surface triangulation, which can be 3D-printed in the highest possible quality. Experiments of this paper are carried out on the dataset of AutoImplant 2021 (https://autoimplant2021.grand-challenge.org/), a MICCAI challenge on cranial implant design. The dataset contains high-resolution skulls that can be viewed as 2D manifolds embedded in a 3D space. Codes associated with this study can be accessed at https://github.com/Jianningli/voxel_rearrangement.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge