Alejandro F. Frangi
Centre for Computational Imaging and Simulation Technologies in Biomedicine
Metrics that matter: Evaluating image quality metrics for medical image generation
May 12, 2025Abstract:Evaluating generative models for synthetic medical imaging is crucial yet challenging, especially given the high standards of fidelity, anatomical accuracy, and safety required for clinical applications. Standard evaluation of generated images often relies on no-reference image quality metrics when ground truth images are unavailable, but their reliability in this complex domain is not well established. This study comprehensively assesses commonly used no-reference image quality metrics using brain MRI data, including tumour and vascular images, providing a representative exemplar for the field. We systematically evaluate metric sensitivity to a range of challenges, including noise, distribution shifts, and, critically, localised morphological alterations designed to mimic clinically relevant inaccuracies. We then compare these metric scores against model performance on a relevant downstream segmentation task, analysing results across both controlled image perturbations and outputs from different generative model architectures. Our findings reveal significant limitations: many widely-used no-reference image quality metrics correlate poorly with downstream task suitability and exhibit a profound insensitivity to localised anatomical details crucial for clinical validity. Furthermore, these metrics can yield misleading scores regarding distribution shifts, e.g. data memorisation. This reveals the risk of misjudging model readiness, potentially leading to the deployment of flawed tools that could compromise patient safety. We conclude that ensuring generative models are truly fit for clinical purpose requires a multifaceted validation framework, integrating performance on relevant downstream tasks with the cautious interpretation of carefully selected no-reference image quality metrics.
From Pixels to Polygons: A Survey of Deep Learning Approaches for Medical Image-to-Mesh Reconstruction
May 06, 2025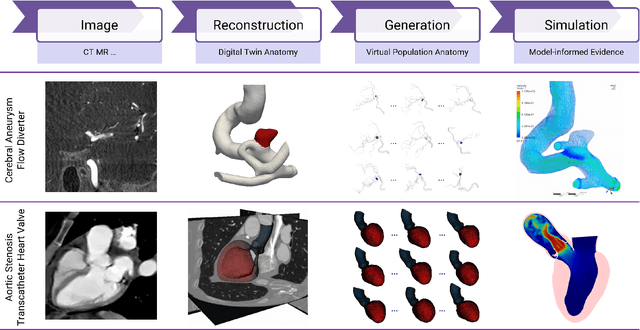
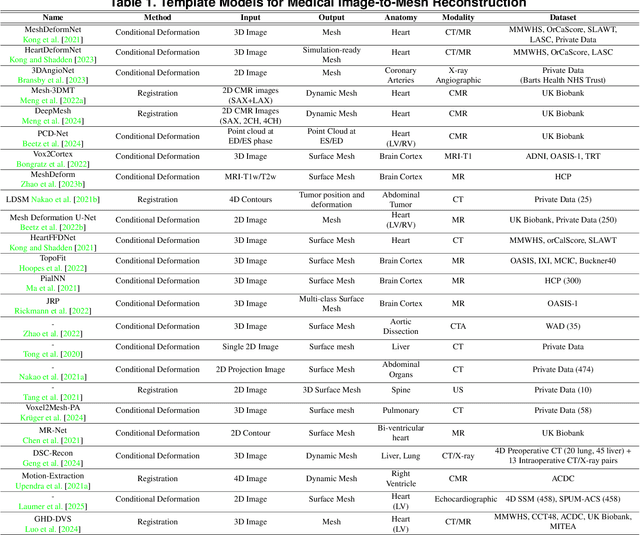
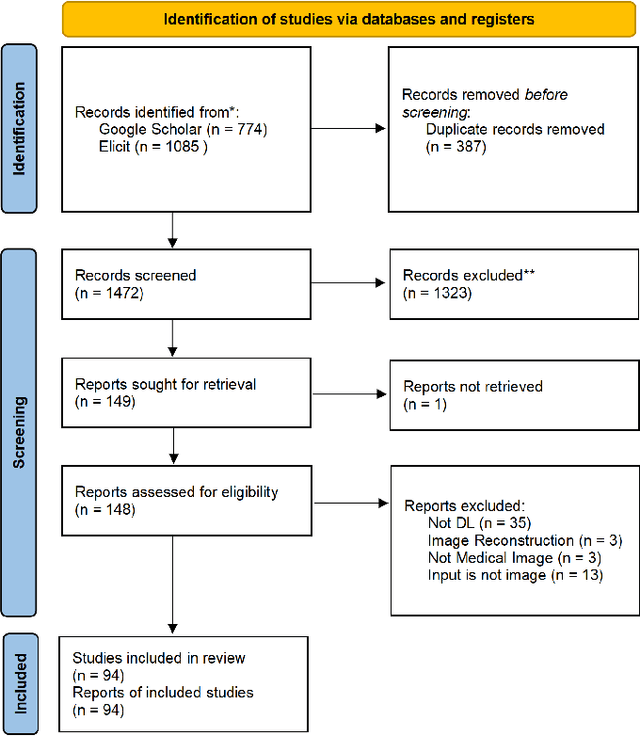
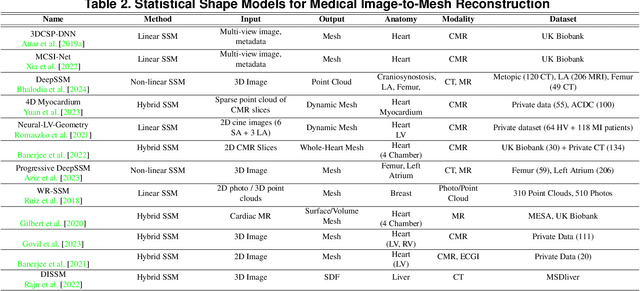
Abstract:Deep learning-based medical image-to-mesh reconstruction has rapidly evolved, enabling the transformation of medical imaging data into three-dimensional mesh models that are critical in computational medicine and in silico trials for advancing our understanding of disease mechanisms, and diagnostic and therapeutic techniques in modern medicine. This survey systematically categorizes existing approaches into four main categories: template models, statistical models, generative models, and implicit models. Each category is analysed in detail, examining their methodological foundations, strengths, limitations, and applicability to different anatomical structures and imaging modalities. We provide an extensive evaluation of these methods across various anatomical applications, from cardiac imaging to neurological studies, supported by quantitative comparisons using standard metrics. Additionally, we compile and analyze major public datasets available for medical mesh reconstruction tasks and discuss commonly used evaluation metrics and loss functions. The survey identifies current challenges in the field, including requirements for topological correctness, geometric accuracy, and multi-modality integration. Finally, we present promising future research directions in this domain. This systematic review aims to serve as a comprehensive reference for researchers and practitioners in medical image analysis and computational medicine.
SACB-Net: Spatial-awareness Convolutions for Medical Image Registration
Mar 25, 2025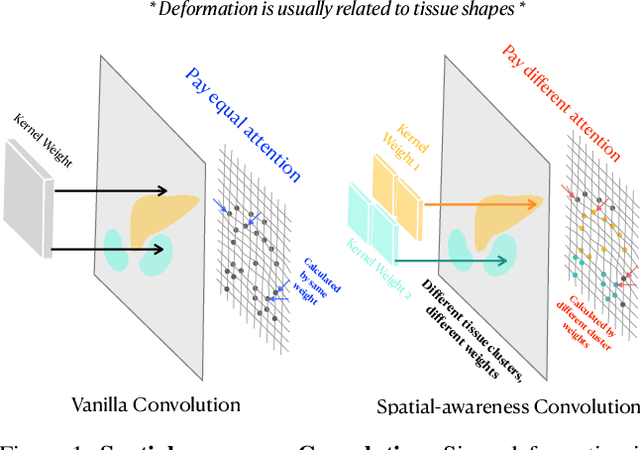
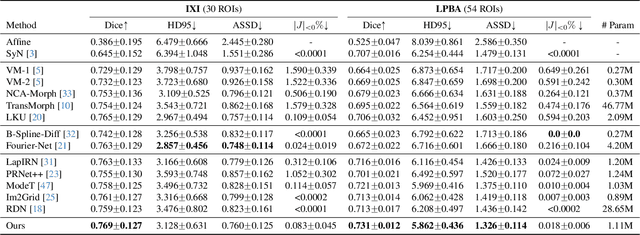
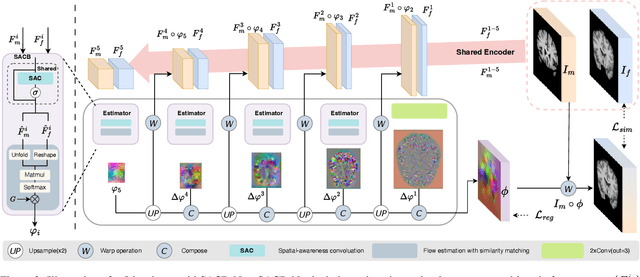
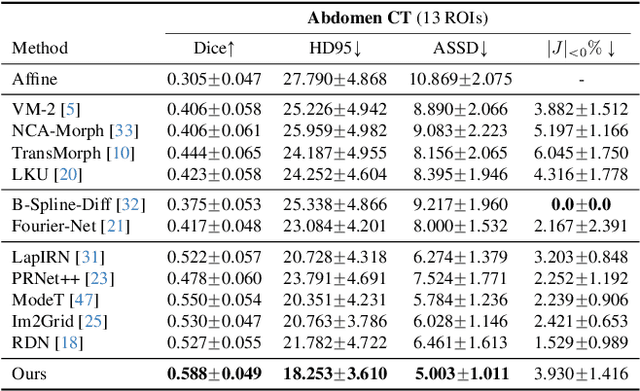
Abstract:Deep learning-based image registration methods have shown state-of-the-art performance and rapid inference speeds. Despite these advances, many existing approaches fall short in capturing spatially varying information in non-local regions of feature maps due to the reliance on spatially-shared convolution kernels. This limitation leads to suboptimal estimation of deformation fields. In this paper, we propose a 3D Spatial-Awareness Convolution Block (SACB) to enhance the spatial information within feature representations. Our SACB estimates the spatial clusters within feature maps by leveraging feature similarity and subsequently parameterizes the adaptive convolution kernels across diverse regions. This adaptive mechanism generates the convolution kernels (weights and biases) tailored to spatial variations, thereby enabling the network to effectively capture spatially varying information. Building on SACB, we introduce a pyramid flow estimator (named SACB-Net) that integrates SACBs to facilitate multi-scale flow composition, particularly addressing large deformations. Experimental results on the brain IXI and LPBA datasets as well as Abdomen CT datasets demonstrate the effectiveness of SACB and the superiority of SACB-Net over the state-of-the-art learning-based registration methods. The code is available at https://github.com/x-xc/SACB_Net .
A Symmetric Dynamic Learning Framework for Diffeomorphic Medical Image Registration
Nov 05, 2024Abstract:Diffeomorphic image registration is crucial for various medical imaging applications because it can preserve the topology of the transformation. This study introduces DCCNN-LSTM-Reg, a learning framework that evolves dynamically and learns a symmetrical registration path by satisfying a specified control increment system. This framework aims to obtain symmetric diffeomorphic deformations between moving and fixed images. To achieve this, we combine deep learning networks with diffeomorphic mathematical mechanisms to create a continuous and dynamic registration architecture, which consists of multiple Symmetric Registration (SR) modules cascaded on five different scales. Specifically, our method first uses two U-nets with shared parameters to extract multiscale feature pyramids from the images. We then develop an SR-module comprising a sequential CNN-LSTM architecture to progressively correct the forward and reverse multiscale deformation fields using control increment learning and the homotopy continuation technique. Through extensive experiments on three 3D registration tasks, we demonstrate that our method outperforms existing approaches in both quantitative and qualitative evaluations.
Statistical Distance-Guided Unsupervised Domain Adaptation for Automated Multi-Class Cardiovascular Magnetic Resonance Image Quality Assessment
Aug 31, 2024Abstract:This study proposes an attention-based statistical distance-guided unsupervised domain adaptation model for multi-class cardiovascular magnetic resonance (CMR) image quality assessment. The proposed model consists of a feature extractor, a label predictor and a statistical distance estimator. An annotated dataset as the source set and an unlabeled dataset as the target set with different statistical distributions are considered inputs. The statistical distance estimator approximates the Wasserstein distance between the extracted feature vectors from the source and target data in a mini-batch. The label predictor predicts data labels of source data and uses a combinational loss function for training, which includes cross entropy and centre loss functions plus the estimated value of the distance estimator. Four datasets, including imaging and k-space data, were used to evaluate the proposed model in identifying four common CMR imaging artefacts: respiratory and cardiac motions, Gibbs ringing and Aliasing. The results of the extensive experiments showed that the proposed model, both in image and k-space analysis, has an acceptable performance in covering the domain shift between the source and target sets. The model explainability evaluations and the ablation studies confirmed the proper functioning and effectiveness of all the model's modules. The proposed model outperformed the previous studies regarding performance and the number of examined artefacts. The proposed model can be used for CMR post-imaging quality control or in large-scale cohort studies for image and k-space quality assessment due to the appropriate performance in domain shift coverage without a tedious data-labelling process.
Multiple Teachers-Meticulous Student: A Domain Adaptive Meta-Knowledge Distillation Model for Medical Image Classification
Mar 17, 2024



Abstract:Background: Image classification can be considered one of the key pillars of medical image analysis. Deep learning (DL) faces challenges that prevent its practical applications despite the remarkable improvement in medical image classification. The data distribution differences can lead to a drop in the efficiency of DL, known as the domain shift problem. Besides, requiring bulk annotated data for model training, the large size of models, and the privacy-preserving of patients are other challenges of using DL in medical image classification. This study presents a strategy that can address the mentioned issues simultaneously. Method: The proposed domain adaptive model based on knowledge distillation can classify images by receiving limited annotated data of different distributions. The designed multiple teachers-meticulous student model trains a student network that tries to solve the challenges by receiving the parameters of several teacher networks. The proposed model was evaluated using six available datasets of different distributions by defining the respiratory motion artefact detection task. Results: The results of extensive experiments using several datasets show the superiority of the proposed model in addressing the domain shift problem and lack of access to bulk annotated data. Besides, the privacy preservation of patients by receiving only the teacher network parameters instead of the original data and consolidating the knowledge of several DL models into a model with almost similar performance are other advantages of the proposed model. Conclusions: The proposed model can pave the way for practical clinical applications of deep classification methods by achieving the mentioned objectives simultaneously.
An End-to-End Deep Learning Generative Framework for Refinable Shape Matching and Generation
Mar 10, 2024



Abstract:Generative modelling for shapes is a prerequisite for In-Silico Clinical Trials (ISCTs), which aim to cost-effectively validate medical device interventions using synthetic anatomical shapes, often represented as 3D surface meshes. However, constructing AI models to generate shapes closely resembling the real mesh samples is challenging due to variable vertex counts, connectivities, and the lack of dense vertex-wise correspondences across the training data. Employing graph representations for meshes, we develop a novel unsupervised geometric deep-learning model to establish refinable shape correspondences in a latent space, construct a population-derived atlas and generate realistic synthetic shapes. We additionally extend our proposed base model to a joint shape generative-clustering multi-atlas framework to incorporate further variability and preserve more details in the generated shapes. Experimental results using liver and left-ventricular models demonstrate the approach's applicability to computational medicine, highlighting its suitability for ISCTs through a comparative analysis.
Unsupervised Domain Adaptation for Brain Vessel Segmentation through Transwarp Contrastive Learning
Feb 23, 2024



Abstract:Unsupervised domain adaptation (UDA) aims to align the labelled source distribution with the unlabelled target distribution to obtain domain-invariant predictive models. Since cross-modality medical data exhibit significant intra and inter-domain shifts and most are unlabelled, UDA is more important while challenging in medical image analysis. This paper proposes a simple yet potent contrastive learning framework for UDA to narrow the inter-domain gap between labelled source and unlabelled target distribution. Our method is validated on cerebral vessel datasets. Experimental results show that our approach can learn latent features from labelled 3DRA modality data and improve vessel segmentation performance in unlabelled MRA modality data.
GS-EMA: Integrating Gradient Surgery Exponential Moving Average with Boundary-Aware Contrastive Learning for Enhanced Domain Generalization in Aneurysm Segmentation
Feb 23, 2024



Abstract:The automated segmentation of cerebral aneurysms is pivotal for accurate diagnosis and treatment planning. Confronted with significant domain shifts and class imbalance in 3D Rotational Angiography (3DRA) data from various medical institutions, the task becomes challenging. These shifts include differences in image appearance, intensity distribution, resolution, and aneurysm size, all of which complicate the segmentation process. To tackle these issues, we propose a novel domain generalization strategy that employs gradient surgery exponential moving average (GS-EMA) optimization technique coupled with boundary-aware contrastive learning (BACL). Our approach is distinct in its ability to adapt to new, unseen domains by learning domain-invariant features, thereby improving the robustness and accuracy of aneurysm segmentation across diverse clinical datasets. The results demonstrate that our proposed approach can extract more domain-invariant features, minimizing over-segmentation and capturing more complete aneurysm structures.
Multi-view Hybrid Graph Convolutional Network for Volume-to-mesh Reconstruction in Cardiovascular MRI
Nov 22, 2023Abstract:Cardiovascular magnetic resonance imaging is emerging as a crucial tool to examine cardiac morphology and function. Essential to this endeavour are anatomical 3D surface and volumetric meshes derived from CMR images, which facilitate computational anatomy studies, biomarker discovery, and in-silico simulations. However, conventional surface mesh generation methods, such as active shape models and multi-atlas segmentation, are highly time-consuming and require complex processing pipelines to generate simulation-ready 3D meshes. In response, we introduce HybridVNet, a novel architecture for direct image-to-mesh extraction seamlessly integrating standard convolutional neural networks with graph convolutions, which we prove can efficiently handle surface and volumetric meshes by encoding them as graph structures. To further enhance accuracy, we propose a multiview HybridVNet architecture which processes both long axis and short axis CMR, showing that it can increase the performance of cardiac MR mesh generation. Our model combines traditional convolutional networks with variational graph generative models, deep supervision and mesh-specific regularisation. Experiments on a comprehensive dataset from the UK Biobank confirm the potential of HybridVNet to significantly advance cardiac imaging and computational cardiology by efficiently generating high-fidelity and simulation ready meshes from CMR images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge