Zeike A. Taylor
Deep-Motion-Net: GNN-based volumetric organ shape reconstruction from single-view 2D projections
Jul 09, 2024



Abstract:We propose Deep-Motion-Net: an end-to-end graph neural network (GNN) architecture that enables 3D (volumetric) organ shape reconstruction from a single in-treatment kV planar X-ray image acquired at any arbitrary projection angle. Estimating and compensating for true anatomical motion during radiotherapy is essential for improving the delivery of planned radiation dose to target volumes while sparing organs-at-risk, and thereby improving the therapeutic ratio. Achieving this using only limited imaging available during irradiation and without the use of surrogate signals or invasive fiducial markers is attractive. The proposed model learns the mesh regression from a patient-specific template and deep features extracted from kV images at arbitrary projection angles. A 2D-CNN encoder extracts image features, and four feature pooling networks fuse these features to the 3D template organ mesh. A ResNet-based graph attention network then deforms the feature-encoded mesh. The model is trained using synthetically generated organ motion instances and corresponding kV images. The latter is generated by deforming a reference CT volume aligned with the template mesh, creating digitally reconstructed radiographs (DRRs) at required projection angles, and DRR-to-kV style transferring with a conditional CycleGAN model. The overall framework was tested quantitatively on synthetic respiratory motion scenarios and qualitatively on in-treatment images acquired over full scan series for liver cancer patients. Overall mean prediction errors for synthetic motion test datasets were 0.16$\pm$0.13 mm, 0.18$\pm$0.19 mm, 0.22$\pm$0.34 mm, and 0.12$\pm$0.11 mm. Mean peak prediction errors were 1.39 mm, 1.99 mm, 3.29 mm, and 1.16 mm.
Biomechanics-informed Non-rigid Medical Image Registration and its Inverse Material Property Estimation with Linear and Nonlinear Elasticity
Jul 03, 2024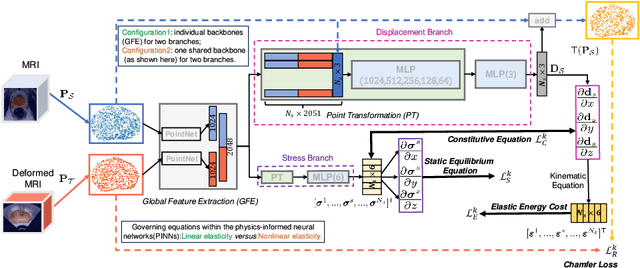
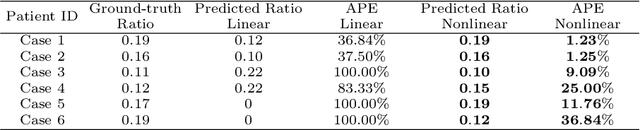
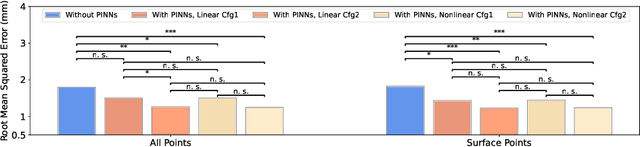
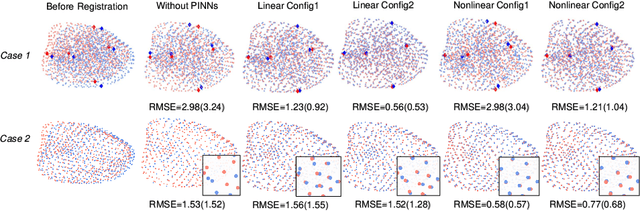
Abstract:This paper investigates both biomechanical-constrained non-rigid medical image registrations and accurate identifications of material properties for soft tissues, using physics-informed neural networks (PINNs). The complex nonlinear elasticity theory is leveraged to formally establish the partial differential equations (PDEs) representing physics laws of biomechanical constraints that need to be satisfied, with which registration and identification tasks are treated as forward (i.e., data-driven solutions of PDEs) and inverse (i.e., parameter estimation) problems under PINNs respectively. Two net configurations (i.e., Cfg1 and Cfg2) have also been compared for both linear and nonlinear physics model. Two sets of experiments have been conducted, using pairs of undeformed and deformed MR images from clinical cases of prostate cancer biopsy. Our contributions are summarised as follows. 1) We developed a learning-based biomechanical-constrained non-rigid registration algorithm using PINNs, where linear elasticity is generalised to the nonlinear version. 2) We demonstrated extensively that nonlinear elasticity shows no statistical significance against linear models in computing point-wise displacement vectors but their respective benefits may depend on specific patients, with finite-element (FE) computed ground-truth. 3) We formulated and solved the inverse parameter estimation problem, under the joint optimisation scheme of registration and parameter identification using PINNs, whose solutions can be accurately found by locating saddle points.
Multi-view Hybrid Graph Convolutional Network for Volume-to-mesh Reconstruction in Cardiovascular MRI
Nov 22, 2023Abstract:Cardiovascular magnetic resonance imaging is emerging as a crucial tool to examine cardiac morphology and function. Essential to this endeavour are anatomical 3D surface and volumetric meshes derived from CMR images, which facilitate computational anatomy studies, biomarker discovery, and in-silico simulations. However, conventional surface mesh generation methods, such as active shape models and multi-atlas segmentation, are highly time-consuming and require complex processing pipelines to generate simulation-ready 3D meshes. In response, we introduce HybridVNet, a novel architecture for direct image-to-mesh extraction seamlessly integrating standard convolutional neural networks with graph convolutions, which we prove can efficiently handle surface and volumetric meshes by encoding them as graph structures. To further enhance accuracy, we propose a multiview HybridVNet architecture which processes both long axis and short axis CMR, showing that it can increase the performance of cardiac MR mesh generation. Our model combines traditional convolutional networks with variational graph generative models, deep supervision and mesh-specific regularisation. Experiments on a comprehensive dataset from the UK Biobank confirm the potential of HybridVNet to significantly advance cardiac imaging and computational cardiology by efficiently generating high-fidelity and simulation ready meshes from CMR images.
Non-rigid Medical Image Registration using Physics-informed Neural Networks
Feb 20, 2023



Abstract:Biomechanical modelling of soft tissue provides a non-data-driven method for constraining medical image registration, such that the estimated spatial transformation is considered biophysically plausible. This has not only been adopted in real-world clinical applications, such as the MR-to-ultrasound registration for prostate intervention of interest in this work, but also provides an explainable means of understanding the organ motion and spatial correspondence establishment. This work instantiates the recently-proposed physics-informed neural networks (PINNs) to a 3D linear elastic model for modelling prostate motion commonly encountered during transrectal ultrasound guided procedures. To overcome a widely-recognised challenge in generalising PINNs to different subjects, we propose to use PointNet as the nodal-permutation-invariant feature extractor, together with a registration algorithm that aligns point sets and simultaneously takes into account the PINN-imposed biomechanics. The proposed method has been both developed and validated in both patient-specific and multi-patient manner.
Prostate motion modelling using biomechanically-trained deep neural networks on unstructured nodes
Jul 09, 2020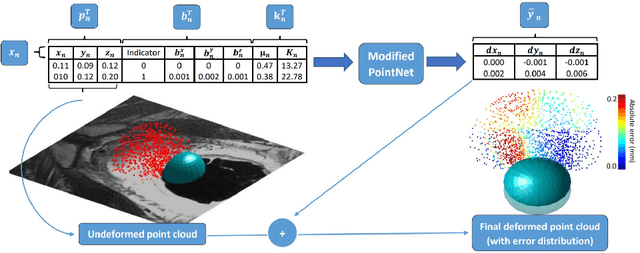
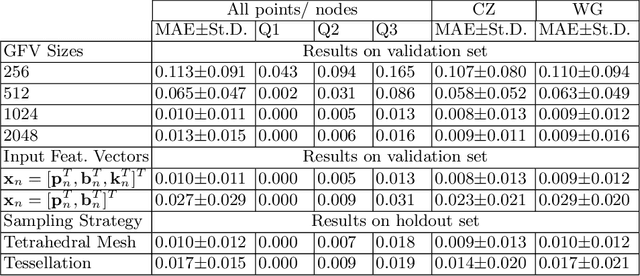
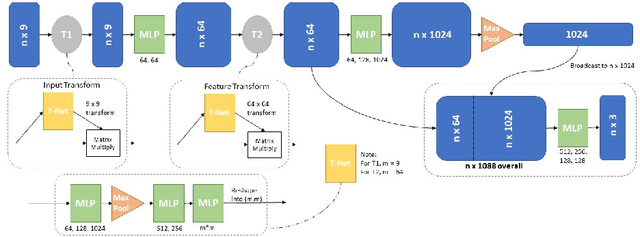
Abstract:In this paper, we propose to train deep neural networks with biomechanical simulations, to predict the prostate motion encountered during ultrasound-guided interventions. In this application, unstructured points are sampled from segmented pre-operative MR images to represent the anatomical regions of interest. The point sets are then assigned with point-specific material properties and displacement loads, forming the un-ordered input feature vectors. An adapted PointNet can be trained to predict the nodal displacements, using finite element (FE) simulations as ground-truth data. Furthermore, a versatile bootstrap aggregating mechanism is validated to accommodate the variable number of feature vectors due to different patient geometries, comprised of a training-time bootstrap sampling and a model averaging inference. This results in a fast and accurate approximation to the FE solutions without requiring subject-specific solid meshing. Based on 160,000 nonlinear FE simulations on clinical imaging data from 320 patients, we demonstrate that the trained networks generalise to unstructured point sets sampled directly from holdout patient segmentation, yielding a near real-time inference and an expected error of 0.017 mm in predicted nodal displacement.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge