Enzo Ferrante
CVN, CentraleSupelec-INRIA, Universite Paris-Saclay, France
CheXmask-U: Quantifying uncertainty in landmark-based anatomical segmentation for X-ray images
Dec 11, 2025Abstract:Uncertainty estimation is essential for the safe clinical deployment of medical image segmentation systems, enabling the identification of unreliable predictions and supporting human oversight. While prior work has largely focused on pixel-level uncertainty, landmark-based segmentation offers inherent topological guarantees yet remains underexplored from an uncertainty perspective. In this work, we study uncertainty estimation for anatomical landmark-based segmentation on chest X-rays. Inspired by hybrid neural network architectures that combine standard image convolutional encoders with graph-based generative decoders, and leveraging their variational latent space, we derive two complementary measures: (i) latent uncertainty, captured directly from the learned distribution parameters, and (ii) predictive uncertainty, obtained by generating multiple stochastic output predictions from latent samples. Through controlled corruption experiments we show that both uncertainty measures increase with perturbation severity, reflecting both global and local degradation. We demonstrate that these uncertainty signals can identify unreliable predictions by comparing with manual ground-truth, and support out-of-distribution detection on the CheXmask dataset. More importantly, we release CheXmask-U (huggingface.co/datasets/mcosarinsky/CheXmask-U), a large scale dataset of 657,566 chest X-ray landmark segmentations with per-node uncertainty estimates, enabling researchers to account for spatial variations in segmentation quality when using these anatomical masks. Our findings establish uncertainty estimation as a promising direction to enhance robustness and safe deployment of landmark-based anatomical segmentation methods in chest X-ray. A fully working interactive demo of the method is available at huggingface.co/spaces/matiasky/CheXmask-U and the source code at github.com/mcosarinsky/CheXmask-U.
PVeRA: Probabilistic Vector-Based Random Matrix Adaptation
Dec 08, 2025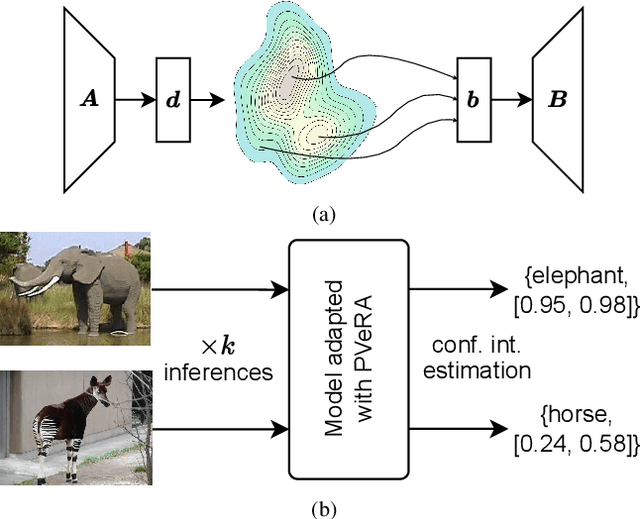
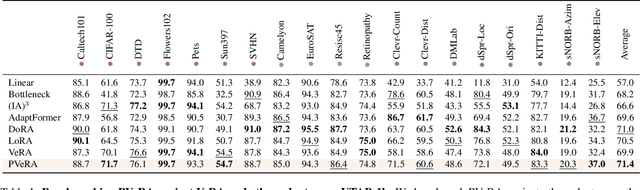
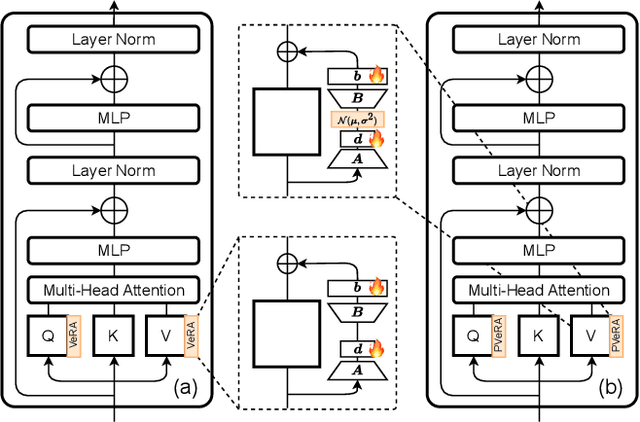

Abstract:Large foundation models have emerged in the last years and are pushing performance boundaries for a variety of tasks. Training or even finetuning such models demands vast datasets and computational resources, which are often scarce and costly. Adaptation methods provide a computationally efficient solution to address these limitations by allowing such models to be finetuned on small amounts of data and computing power. This is achieved by appending new trainable modules to frozen backbones with only a fraction of the trainable parameters and fitting only these modules on novel tasks. Recently, the VeRA adapter was shown to excel in parameter-efficient adaptations by utilizing a pair of frozen random low-rank matrices shared across all layers. In this paper, we propose PVeRA, a probabilistic version of the VeRA adapter, which modifies the low-rank matrices of VeRA in a probabilistic manner. This modification naturally allows handling inherent ambiguities in the input and allows for different sampling configurations during training and testing. A comprehensive evaluation was performed on the VTAB-1k benchmark and seven adapters, with PVeRA outperforming VeRA and other adapters. Our code for training models with PVeRA and benchmarking all adapters is available https://github.com/leofillioux/pvera.
On the Risk of Misleading Reports: Diagnosing Textual Biases in Multimodal Clinical AI
Jul 31, 2025



Abstract:Clinical decision-making relies on the integrated analysis of medical images and the associated clinical reports. While Vision-Language Models (VLMs) can offer a unified framework for such tasks, they can exhibit strong biases toward one modality, frequently overlooking critical visual cues in favor of textual information. In this work, we introduce Selective Modality Shifting (SMS), a perturbation-based approach to quantify a model's reliance on each modality in binary classification tasks. By systematically swapping images or text between samples with opposing labels, we expose modality-specific biases. We assess six open-source VLMs-four generalist models and two fine-tuned for medical data-on two medical imaging datasets with distinct modalities: MIMIC-CXR (chest X-ray) and FairVLMed (scanning laser ophthalmoscopy). By assessing model performance and the calibration of every model in both unperturbed and perturbed settings, we reveal a marked dependency on text input, which persists despite the presence of complementary visual information. We also perform a qualitative attention-based analysis which further confirms that image content is often overshadowed by text details. Our findings highlight the importance of designing and evaluating multimodal medical models that genuinely integrate visual and textual cues, rather than relying on single-modality signals.
Towards Reliable WMH Segmentation under Domain Shift: An Application Study using Maximum Entropy Regularization to Improve Uncertainty Estimation
Jun 17, 2025Abstract:Accurate segmentation of white matter hyperintensities (WMH) is crucial for clinical decision-making, particularly in the context of multiple sclerosis. However, domain shifts, such as variations in MRI machine types or acquisition parameters, pose significant challenges to model calibration and uncertainty estimation. This study investigates the impact of domain shift on WMH segmentation by proposing maximum-entropy regularization techniques to enhance model calibration and uncertainty estimation, with the purpose of identifying errors post-deployment using predictive uncertainty as a proxy measure that does not require ground-truth labels. To do this, we conducted experiments using a U-Net architecture to evaluate these regularization schemes on two publicly available datasets, assessing performance with the Dice coefficient, expected calibration error, and entropy-based uncertainty estimates. Our results show that entropy-based uncertainty estimates can anticipate segmentation errors, and that maximum-entropy regularization further strengthens the correlation between uncertainty and segmentation performance while also improving model calibration under domain shift.
Performance Estimation for Supervised Medical Image Segmentation Models on Unlabeled Data Using UniverSeg
Apr 22, 2025Abstract:The performance of medical image segmentation models is usually evaluated using metrics like the Dice score and Hausdorff distance, which compare predicted masks to ground truth annotations. However, when applying the model to unseen data, such as in clinical settings, it is often impractical to annotate all the data, making the model's performance uncertain. To address this challenge, we propose the Segmentation Performance Evaluator (SPE), a framework for estimating segmentation models' performance on unlabeled data. This framework is adaptable to various evaluation metrics and model architectures. Experiments on six publicly available datasets across six evaluation metrics including pixel-based metrics such as Dice score and distance-based metrics like HD95, demonstrated the versatility and effectiveness of our approach, achieving a high correlation (0.956$\pm$0.046) and low MAE (0.025$\pm$0.019) compare with real Dice score on the independent test set. These results highlight its ability to reliably estimate model performance without requiring annotations. The SPE framework integrates seamlessly into any model training process without adding training overhead, enabling performance estimation and facilitating the real-world application of medical image segmentation algorithms. The source code is publicly available
ChronoRoot 2.0: An Open AI-Powered Platform for 2D Temporal Plant Phenotyping
Apr 20, 2025



Abstract:The analysis of plant developmental plasticity, including root system architecture, is fundamental to understanding plant adaptability and development, particularly in the context of climate change and agricultural sustainability. While significant advances have been made in plant phenotyping technologies, comprehensive temporal analysis of root development remains challenging, with most existing solutions providing either limited throughput or restricted structural analysis capabilities. Here, we present ChronoRoot 2.0, an integrated open-source platform that combines affordable hardware with advanced artificial intelligence to enable sophisticated temporal plant phenotyping. The system introduces several major advances, offering an integral perspective of seedling development: (i) simultaneous multi-organ tracking of six distinct plant structures, (ii) quality control through real-time validation, (iii) comprehensive architectural measurements including novel gravitropic response parameters, and (iv) dual specialized user interfaces for both architectural analysis and high-throughput screening. We demonstrate the system's capabilities through three use cases for Arabidopsis thaliana: characterization of circadian growth patterns under different light conditions, detailed analysis of gravitropic responses in transgenic plants, and high-throughput screening of etiolation responses across multiple genotypes. ChronoRoot 2.0 maintains its predecessor's advantages of low cost and modularity while significantly expanding its capabilities, making sophisticated temporal phenotyping more accessible to the broader plant science community. The system's open-source nature, combined with extensive documentation and containerized deployment options, ensures reproducibility and enables community-driven development of new analytical capabilities.
Kaleidoscope: In-language Exams for Massively Multilingual Vision Evaluation
Apr 09, 2025Abstract:The evaluation of vision-language models (VLMs) has mainly relied on English-language benchmarks, leaving significant gaps in both multilingual and multicultural coverage. While multilingual benchmarks have expanded, both in size and languages, many rely on translations of English datasets, failing to capture cultural nuances. In this work, we propose Kaleidoscope, as the most comprehensive exam benchmark to date for the multilingual evaluation of vision-language models. Kaleidoscope is a large-scale, in-language multimodal benchmark designed to evaluate VLMs across diverse languages and visual inputs. Kaleidoscope covers 18 languages and 14 different subjects, amounting to a total of 20,911 multiple-choice questions. Built through an open science collaboration with a diverse group of researchers worldwide, Kaleidoscope ensures linguistic and cultural authenticity. We evaluate top-performing multilingual vision-language models and find that they perform poorly on low-resource languages and in complex multimodal scenarios. Our results highlight the need for progress on culturally inclusive multimodal evaluation frameworks.
In-Context Reverse Classification Accuracy: Efficient Estimation of Segmentation Quality without Ground-Truth
Mar 06, 2025Abstract:Assessing the quality of automatic image segmentation is crucial in clinical practice, but often very challenging due to the limited availability of ground truth annotations. In this paper, we introduce In-Context Reverse Classification Accuracy (In-Context RCA), a novel framework for automatically estimating segmentation quality in the absence of ground-truth annotations. By leveraging recent in-context learning segmentation models and incorporating retrieval-augmentation techniques to select the most relevant reference images, our approach enables efficient quality estimation with minimal reference data. Validated across diverse medical imaging modalities, our method demonstrates robust performance and computational efficiency, offering a promising solution for automated quality control in clinical workflows, where fast and reliable segmentation assessment is essential. The code is available at https://github.com/mcosarinsky/In-Context-RCA.
Fairness of Deep Ensembles: On the interplay between per-group task difficulty and under-representation
Jan 24, 2025



Abstract:Ensembling is commonly regarded as an effective way to improve the general performance of models in machine learning, while also increasing the robustness of predictions. When it comes to algorithmic fairness, heterogeneous ensembles, composed of multiple model types, have been employed to mitigate biases in terms of demographic attributes such as sex, age or ethnicity. Moreover, recent work has shown how in multi-class problems even simple homogeneous ensembles may favor performance of the worst-performing target classes. While homogeneous ensembles are simpler to implement in practice, it is not yet clear whether their benefits translate to groups defined not in terms of their target class, but in terms of demographic or protected attributes, hence improving fairness. In this work we show how this simple and straightforward method is indeed able to mitigate disparities, particularly benefiting under-performing subgroups. Interestingly, this can be achieved without sacrificing overall performance, which is a common trade-off observed in bias mitigation strategies. Moreover, we analyzed the interplay between two factors which may result in biases: sub-group under-representation and the inherent difficulty of the task for each group. These results revealed that, contrary to popular assumptions, having balanced datasets may be suboptimal if the task difficulty varies between subgroups. Indeed, we found that a perfectly balanced dataset may hurt both the overall performance and the gap between groups. This highlights the importance of considering the interaction between multiple forces at play in fairness.
In the Picture: Medical Imaging Datasets, Artifacts, and their Living Review
Jan 18, 2025


Abstract:Datasets play a critical role in medical imaging research, yet issues such as label quality, shortcuts, and metadata are often overlooked. This lack of attention may harm the generalizability of algorithms and, consequently, negatively impact patient outcomes. While existing medical imaging literature reviews mostly focus on machine learning (ML) methods, with only a few focusing on datasets for specific applications, these reviews remain static -- they are published once and not updated thereafter. This fails to account for emerging evidence, such as biases, shortcuts, and additional annotations that other researchers may contribute after the dataset is published. We refer to these newly discovered findings of datasets as research artifacts. To address this gap, we propose a living review that continuously tracks public datasets and their associated research artifacts across multiple medical imaging applications. Our approach includes a framework for the living review to monitor data documentation artifacts, and an SQL database to visualize the citation relationships between research artifact and dataset. Lastly, we discuss key considerations for creating medical imaging datasets, review best practices for data annotation, discuss the significance of shortcuts and demographic diversity, and emphasize the importance of managing datasets throughout their entire lifecycle. Our demo is publicly available at http://130.226.140.142.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge