Dovile Juodelyte
Learning to Harmonize Cross-vendor X-ray Images by Non-linear Image Dynamics Correction
Apr 14, 2025Abstract:In this paper, we explore how conventional image enhancement can improve model robustness in medical image analysis. By applying commonly used normalization methods to images from various vendors and studying their influence on model generalization in transfer learning, we show that the nonlinear characteristics of domain-specific image dynamics cannot be addressed by simple linear transforms. To tackle this issue, we reformulate the image harmonization task as an exposure correction problem and propose a method termed Global Deep Curve Estimation (GDCE) to reduce domain-specific exposure mismatch. GDCE performs enhancement via a pre-defined polynomial function and is trained with the help of a ``domain discriminator'', aiming to improve model transparency in downstream tasks compared to existing black-box methods.
In the Picture: Medical Imaging Datasets, Artifacts, and their Living Review
Jan 18, 2025


Abstract:Datasets play a critical role in medical imaging research, yet issues such as label quality, shortcuts, and metadata are often overlooked. This lack of attention may harm the generalizability of algorithms and, consequently, negatively impact patient outcomes. While existing medical imaging literature reviews mostly focus on machine learning (ML) methods, with only a few focusing on datasets for specific applications, these reviews remain static -- they are published once and not updated thereafter. This fails to account for emerging evidence, such as biases, shortcuts, and additional annotations that other researchers may contribute after the dataset is published. We refer to these newly discovered findings of datasets as research artifacts. To address this gap, we propose a living review that continuously tracks public datasets and their associated research artifacts across multiple medical imaging applications. Our approach includes a framework for the living review to monitor data documentation artifacts, and an SQL database to visualize the citation relationships between research artifact and dataset. Lastly, we discuss key considerations for creating medical imaging datasets, review best practices for data annotation, discuss the significance of shortcuts and demographic diversity, and emphasize the importance of managing datasets throughout their entire lifecycle. Our demo is publicly available at http://130.226.140.142.
On dataset transferability in medical image classification
Dec 28, 2024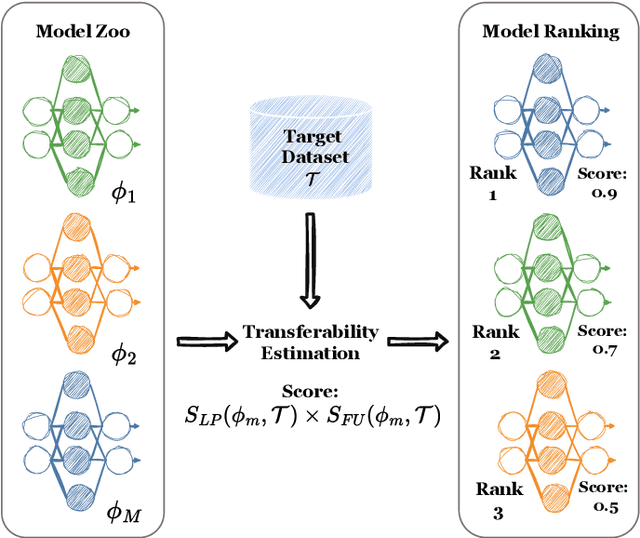
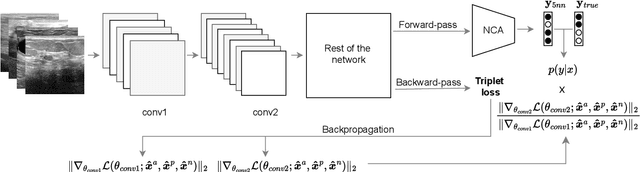
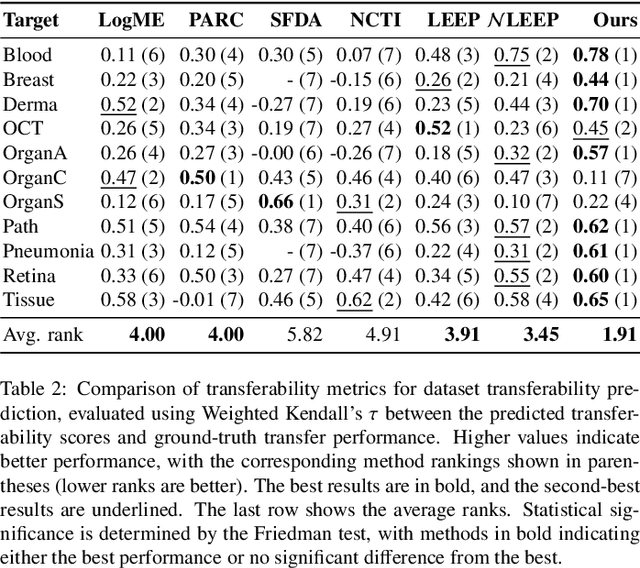
Abstract:Current transferability estimation methods designed for natural image datasets are often suboptimal in medical image classification. These methods primarily focus on estimating the suitability of pre-trained source model features for a target dataset, which can lead to unrealistic predictions, such as suggesting that the target dataset is the best source for itself. To address this, we propose a novel transferability metric that combines feature quality with gradients to evaluate both the suitability and adaptability of source model features for target tasks. We evaluate our approach in two new scenarios: source dataset transferability for medical image classification and cross-domain transferability. Our results show that our method outperforms existing transferability metrics in both settings. We also provide insight into the factors influencing transfer performance in medical image classification, as well as the dynamics of cross-domain transfer from natural to medical images. Additionally, we provide ground-truth transfer performance benchmarking results to encourage further research into transferability estimation for medical image classification. Our code and experiments are available at https://github.com/DovileDo/transferability-in-medical-imaging.
Exploring connections of spectral analysis and transfer learning in medical imaging
Jul 16, 2024



Abstract:In this paper, we use spectral analysis to investigate transfer learning and study model sensitivity to frequency shortcuts in medical imaging. By analyzing the power spectrum density of both pre-trained and fine-tuned model gradients, as well as artificially generated frequency shortcuts, we observe notable differences in learning priorities between models pre-trained on natural vs medical images, which generally persist during fine-tuning. We find that when a model's learning priority aligns with the power spectrum density of an artifact, it results in overfitting to that artifact. Based on these observations, we show that source data editing can alter the model's resistance to shortcut learning.
Source Matters: Source Dataset Impact on Model Robustness in Medical Imaging
Mar 07, 2024Abstract:Transfer learning has become an essential part of medical imaging classification algorithms, often leveraging ImageNet weights. However, the domain shift from natural to medical images has prompted alternatives such as RadImageNet, often demonstrating comparable classification performance. However, it remains unclear whether the performance gains from transfer learning stem from improved generalization or shortcut learning. To address this, we investigate potential confounders -- whether synthetic or sampled from the data -- across two publicly available chest X-ray and CT datasets. We show that ImageNet and RadImageNet achieve comparable classification performance, yet ImageNet is much more prone to overfitting to confounders. We recommend that researchers using ImageNet-pretrained models reexamine their model robustness by conducting similar experiments. Our code and experiments are available at https://github.com/DovileDo/source-matters.
Towards actionability for open medical imaging datasets: lessons from community-contributed platforms for data management and stewardship
Feb 09, 2024



Abstract:Medical imaging datasets are fundamental to artificial intelligence (AI) in healthcare. The accuracy, robustness and fairness of diagnostic algorithms depend on the data (and its quality) on which the models are trained and evaluated. Medical imaging datasets have become increasingly available to the public, and are often hosted on Community-Contributed Platforms (CCP), including private companies like Kaggle or HuggingFace. While open data is important to enhance the redistribution of data's public value, we find that the current CCP governance model fails to uphold the quality needed and recommended practices for sharing, documenting, and evaluating datasets. In this paper we investigate medical imaging datasets on CCPs and how they are documented, shared, and maintained. We first highlight some differences between medical imaging and computer vision, particularly in the potentially harmful downstream effects due to poor adoption of recommended dataset management practices. We then analyze 20 (10 medical and 10 computer vision) popular datasets on CCPs and find vague licenses, lack of persistent identifiers and storage, duplicates and missing metadata, with differences between the platforms. We present "actionability" as a conceptual metric to reveal the data quality gap between characteristics of data on CCPs and the desired characteristics of data for AI in healthcare. Finally, we propose a commons-based stewardship model for documenting, sharing and maintaining datasets on CCPs and end with a discussion of limitations and open questions.
Data usage and citation practices in medical imaging conferences
Feb 05, 2024



Abstract:Medical imaging papers often focus on methodology, but the quality of the algorithms and the validity of the conclusions are highly dependent on the datasets used. As creating datasets requires a lot of effort, researchers often use publicly available datasets, there is however no adopted standard for citing the datasets used in scientific papers, leading to difficulty in tracking dataset usage. In this work, we present two open-source tools we created that could help with the detection of dataset usage, a pipeline \url{https://github.com/TheoSourget/Public_Medical_Datasets_References} using OpenAlex and full-text analysis, and a PDF annotation software \url{https://github.com/TheoSourget/pdf_annotator} used in our study to manually label the presence of datasets. We applied both tools on a study of the usage of 20 publicly available medical datasets in papers from MICCAI and MIDL. We compute the proportion and the evolution between 2013 and 2023 of 3 types of presence in a paper: cited, mentioned in the full text, cited and mentioned. Our findings demonstrate the concentration of the usage of a limited set of datasets. We also highlight different citing practices, making the automation of tracking difficult.
Augmenting Chest X-ray Datasets with Non-Expert Annotations
Sep 05, 2023Abstract:The advancement of machine learning algorithms in medical image analysis requires the expansion of training datasets. A popular and cost-effective approach is automated annotation extraction from free-text medical reports, primarily due to the high costs associated with expert clinicians annotating chest X-ray images. However, it has been shown that the resulting datasets are susceptible to biases and shortcuts. Another strategy to increase the size of a dataset is crowdsourcing, a widely adopted practice in general computer vision with some success in medical image analysis. In a similar vein to crowdsourcing, we enhance two publicly available chest X-ray datasets by incorporating non-expert annotations. However, instead of using diagnostic labels, we annotate shortcuts in the form of tubes. We collect 3.5k chest drain annotations for CXR14, and 1k annotations for 4 different tube types in PadChest. We train a chest drain detector with the non-expert annotations that generalizes well to expert labels. Moreover, we compare our annotations to those provided by experts and show "moderate" to "almost perfect" agreement. Finally, we present a pathology agreement study to raise awareness about ground truth annotations. We make our annotations and code available.
Revisiting Hidden Representations in Transfer Learning for Medical Imaging
Feb 16, 2023


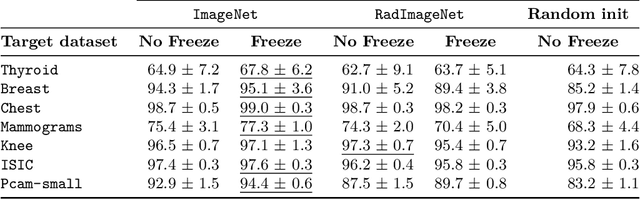
Abstract:While a key component to the success of deep learning is the availability of massive amounts of training data, medical image datasets are often limited in diversity and size. Transfer learning has the potential to bridge the gap between related yet different domains. For medical applications, however, it remains unclear whether it is more beneficial to pre-train on natural or medical images. We aim to shed light on this problem by comparing initialization on ImageNet and RadImageNet on seven medical classification tasks. We investigate their learned representations with Canonical Correlation Analysis (CCA) and compare the predictions of the different models. We find that overall the models pre-trained on ImageNet outperform those trained on RadImageNet. Our results show that, contrary to intuition, ImageNet and RadImageNet converge to distinct intermediate representations, and that these representations are even more dissimilar after fine-tuning. Despite these distinct representations, the predictions of the models remain similar. Our findings challenge the notion that transfer learning is effective due to the reuse of general features in the early layers of a convolutional neural network and show that weight similarity before and after fine-tuning is negatively related to performance gains.
Detecting Shortcuts in Medical Images -- A Case Study in Chest X-rays
Nov 09, 2022Abstract:The availability of large public datasets and the increased amount of computing power have shifted the interest of the medical community to high-performance algorithms. However, little attention is paid to the quality of the data and their annotations. High performance on benchmark datasets may be reported without considering possible shortcuts or artifacts in the data, besides, models are not tested on subpopulation groups. With this work, we aim to raise awareness about shortcuts problems. We validate previous findings, and present a case study on chest X-rays using two publicly available datasets. We share annotations for a subset of pneumothorax images with drains. We conclude with general recommendations for medical image classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge