Théo Sourget
In the Picture: Medical Imaging Datasets, Artifacts, and their Living Review
Jan 18, 2025


Abstract:Datasets play a critical role in medical imaging research, yet issues such as label quality, shortcuts, and metadata are often overlooked. This lack of attention may harm the generalizability of algorithms and, consequently, negatively impact patient outcomes. While existing medical imaging literature reviews mostly focus on machine learning (ML) methods, with only a few focusing on datasets for specific applications, these reviews remain static -- they are published once and not updated thereafter. This fails to account for emerging evidence, such as biases, shortcuts, and additional annotations that other researchers may contribute after the dataset is published. We refer to these newly discovered findings of datasets as research artifacts. To address this gap, we propose a living review that continuously tracks public datasets and their associated research artifacts across multiple medical imaging applications. Our approach includes a framework for the living review to monitor data documentation artifacts, and an SQL database to visualize the citation relationships between research artifact and dataset. Lastly, we discuss key considerations for creating medical imaging datasets, review best practices for data annotation, discuss the significance of shortcuts and demographic diversity, and emphasize the importance of managing datasets throughout their entire lifecycle. Our demo is publicly available at http://130.226.140.142.
Mask of truth: model sensitivity to unexpected regions of medical images
Dec 05, 2024Abstract:The development of larger models for medical image analysis has led to increased performance. However, it also affected our ability to explain and validate model decisions. Models can use non-relevant parts of images, also called spurious correlations or shortcuts, to obtain high performance on benchmark datasets but fail in real-world scenarios. In this work, we challenge the capacity of convolutional neural networks (CNN) to classify chest X-rays and eye fundus images while masking out clinically relevant parts of the image. We show that all models trained on the PadChest dataset, irrespective of the masking strategy, are able to obtain an Area Under the Curve (AUC) above random. Moreover, the models trained on full images obtain good performance on images without the region of interest (ROI), even superior to the one obtained on images only containing the ROI. We also reveal a possible spurious correlation in the Chaksu dataset while the performances are more aligned with the expectation of an unbiased model. We go beyond the performance analysis with the usage of the explainability method SHAP and the analysis of embeddings. We asked a radiology resident to interpret chest X-rays under different masking to complement our findings with clinical knowledge. Our code is available at https://github.com/TheoSourget/MMC_Masking and https://github.com/TheoSourget/MMC_Masking_EyeFundus
Towards actionability for open medical imaging datasets: lessons from community-contributed platforms for data management and stewardship
Feb 09, 2024



Abstract:Medical imaging datasets are fundamental to artificial intelligence (AI) in healthcare. The accuracy, robustness and fairness of diagnostic algorithms depend on the data (and its quality) on which the models are trained and evaluated. Medical imaging datasets have become increasingly available to the public, and are often hosted on Community-Contributed Platforms (CCP), including private companies like Kaggle or HuggingFace. While open data is important to enhance the redistribution of data's public value, we find that the current CCP governance model fails to uphold the quality needed and recommended practices for sharing, documenting, and evaluating datasets. In this paper we investigate medical imaging datasets on CCPs and how they are documented, shared, and maintained. We first highlight some differences between medical imaging and computer vision, particularly in the potentially harmful downstream effects due to poor adoption of recommended dataset management practices. We then analyze 20 (10 medical and 10 computer vision) popular datasets on CCPs and find vague licenses, lack of persistent identifiers and storage, duplicates and missing metadata, with differences between the platforms. We present "actionability" as a conceptual metric to reveal the data quality gap between characteristics of data on CCPs and the desired characteristics of data for AI in healthcare. Finally, we propose a commons-based stewardship model for documenting, sharing and maintaining datasets on CCPs and end with a discussion of limitations and open questions.
Detection Transformer for Teeth Detection, Segmentation, and Numbering in Oral Rare Diseases: Focus on Data Augmentation and Inpainting Techniques
Feb 06, 2024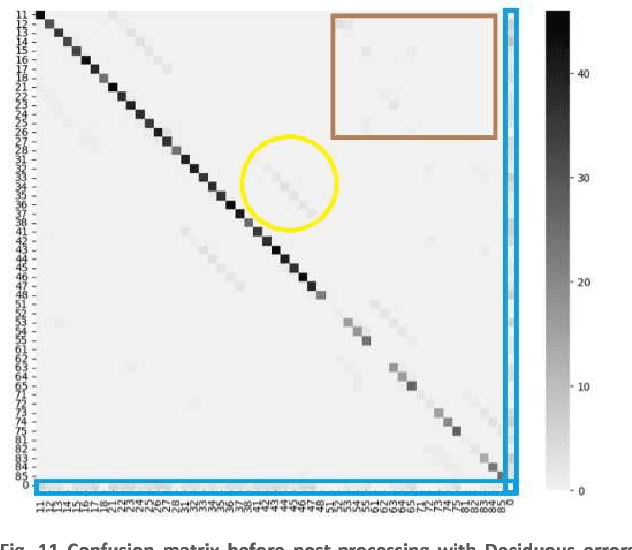
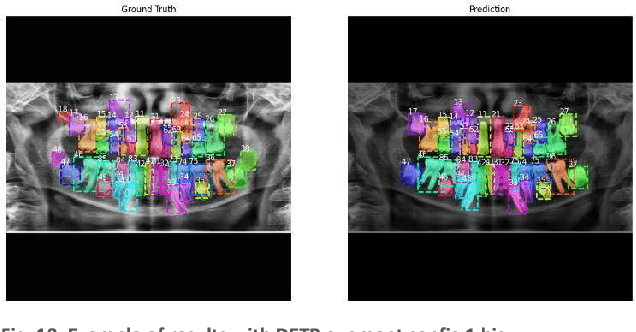
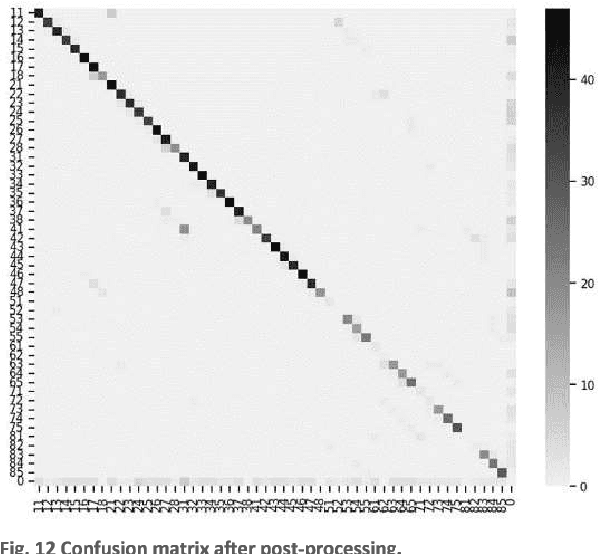
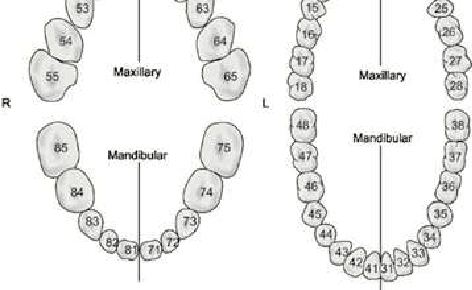
Abstract:In this work, we focused on deep learning image processing in the context of oral rare diseases, which pose challenges due to limited data availability. A crucial step involves teeth detection, segmentation and numbering in panoramic radiographs. To this end, we used a dataset consisting of 156 panoramic radiographs from individuals with rare oral diseases and labeled by experts. We trained the Detection Transformer (DETR) neural network for teeth detection, segmentation, and numbering the 52 teeth classes. In addition, we used data augmentation techniques, including geometric transformations. Finally, we generated new panoramic images using inpainting techniques with stable diffusion, by removing teeth from a panoramic radiograph and integrating teeth into it. The results showed a mAP exceeding 0,69 for DETR without data augmentation. The mAP was improved to 0,82 when data augmentation techniques are used. Furthermore, we observed promising performances when using new panoramic radiographs generated with inpainting technique, with mAP of 0,76.
Data usage and citation practices in medical imaging conferences
Feb 05, 2024



Abstract:Medical imaging papers often focus on methodology, but the quality of the algorithms and the validity of the conclusions are highly dependent on the datasets used. As creating datasets requires a lot of effort, researchers often use publicly available datasets, there is however no adopted standard for citing the datasets used in scientific papers, leading to difficulty in tracking dataset usage. In this work, we present two open-source tools we created that could help with the detection of dataset usage, a pipeline \url{https://github.com/TheoSourget/Public_Medical_Datasets_References} using OpenAlex and full-text analysis, and a PDF annotation software \url{https://github.com/TheoSourget/pdf_annotator} used in our study to manually label the presence of datasets. We applied both tools on a study of the usage of 20 publicly available medical datasets in papers from MICCAI and MIDL. We compute the proportion and the evolution between 2013 and 2023 of 3 types of presence in a paper: cited, mentioned in the full text, cited and mentioned. Our findings demonstrate the concentration of the usage of a limited set of datasets. We also highlight different citing practices, making the automation of tracking difficult.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge