Caroline Petitjean
MiSuRe is all you need to explain your image segmentation
Jun 18, 2024



Abstract:The last decade of computer vision has been dominated by Deep Learning architectures, thanks to their unparalleled success. Their performance, however, often comes at the cost of explainability owing to their highly non-linear nature. Consequently, a parallel field of eXplainable Artificial Intelligence (XAI) has developed with the aim of generating insights regarding the decision making process of deep learning models. An important problem in XAI is that of the generation of saliency maps. These are regions in an input image which contributed most towards the model's final decision. Most work in this regard, however, has been focused on image classification, and image segmentation - despite being a ubiquitous task - has not received the same attention. In the present work, we propose MiSuRe (Minimally Sufficient Region) as an algorithm to generate saliency maps for image segmentation. The goal of the saliency maps generated by MiSuRe is to get rid of irrelevant regions, and only highlight those regions in the input image which are crucial to the image segmentation decision. We perform our analysis on 3 datasets: Triangle (artificially constructed), COCO-2017 (natural images), and the Synapse multi-organ (medical images). Additionally, we identify a potential usecase of these post-hoc saliency maps in order to perform post-hoc reliability of the segmentation model.
Data usage and citation practices in medical imaging conferences
Feb 05, 2024



Abstract:Medical imaging papers often focus on methodology, but the quality of the algorithms and the validity of the conclusions are highly dependent on the datasets used. As creating datasets requires a lot of effort, researchers often use publicly available datasets, there is however no adopted standard for citing the datasets used in scientific papers, leading to difficulty in tracking dataset usage. In this work, we present two open-source tools we created that could help with the detection of dataset usage, a pipeline \url{https://github.com/TheoSourget/Public_Medical_Datasets_References} using OpenAlex and full-text analysis, and a PDF annotation software \url{https://github.com/TheoSourget/pdf_annotator} used in our study to manually label the presence of datasets. We applied both tools on a study of the usage of 20 publicly available medical datasets in papers from MICCAI and MIDL. We compute the proportion and the evolution between 2013 and 2023 of 3 types of presence in a paper: cited, mentioned in the full text, cited and mentioned. Our findings demonstrate the concentration of the usage of a limited set of datasets. We also highlight different citing practices, making the automation of tracking difficult.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Effect of Prior-based Losses on Segmentation Performance: A Benchmark
Jan 12, 2022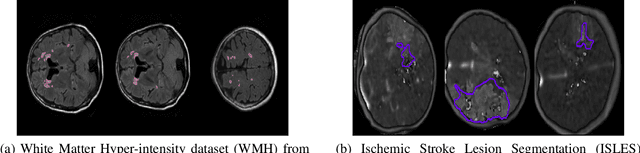
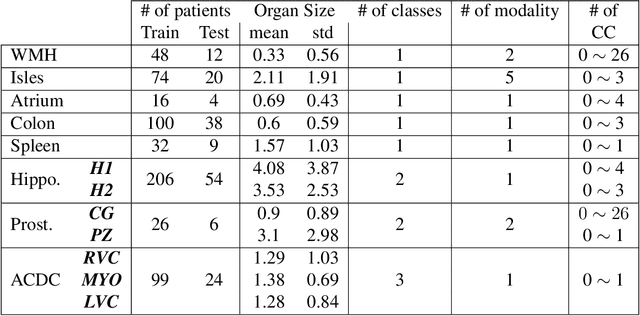
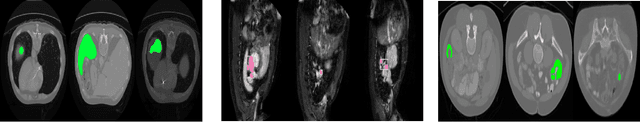

Abstract:Today, deep convolutional neural networks (CNNs) have demonstrated state-of-the-art performance for medical image segmentation, on various imaging modalities and tasks. Despite early success, segmentation networks may still generate anatomically aberrant segmentations, with holes or inaccuracies near the object boundaries. To enforce anatomical plausibility, recent research studies have focused on incorporating prior knowledge such as object shape or boundary, as constraints in the loss function. Prior integrated could be low-level referring to reformulated representations extracted from the ground-truth segmentations, or high-level representing external medical information such as the organ's shape or size. Over the past few years, prior-based losses exhibited a rising interest in the research field since they allow integration of expert knowledge while still being architecture-agnostic. However, given the diversity of prior-based losses on different medical imaging challenges and tasks, it has become hard to identify what loss works best for which dataset. In this paper, we establish a benchmark of recent prior-based losses for medical image segmentation. The main objective is to provide intuition onto which losses to choose given a particular task or dataset. To this end, four low-level and high-level prior-based losses are selected. The considered losses are validated on 8 different datasets from a variety of medical image segmentation challenges including the Decathlon, the ISLES and the WMH challenge. Results show that whereas low-level prior-based losses can guarantee an increase in performance over the Dice loss baseline regardless of the dataset characteristics, high-level prior-based losses can increase anatomical plausibility as per data characteristics.
High-level Prior-based Loss Functions for Medical Image Segmentation: A Survey
Nov 22, 2020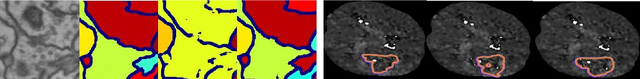
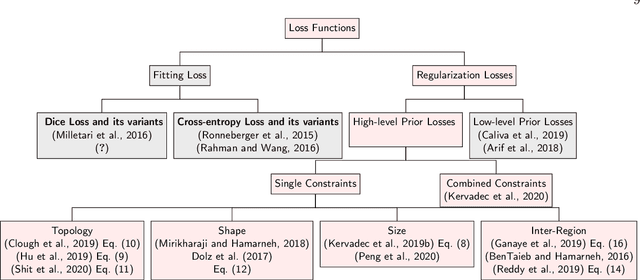
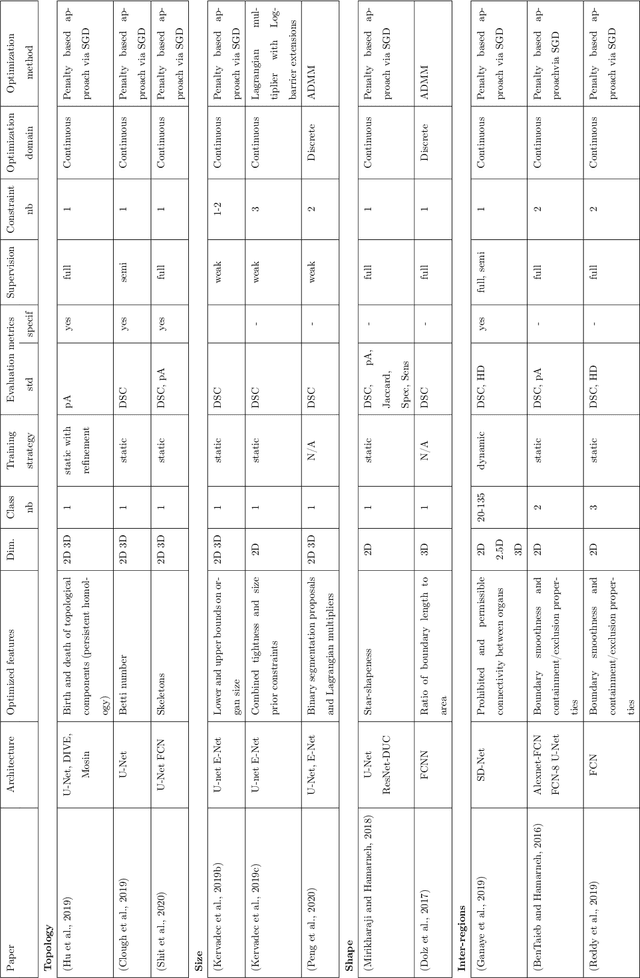
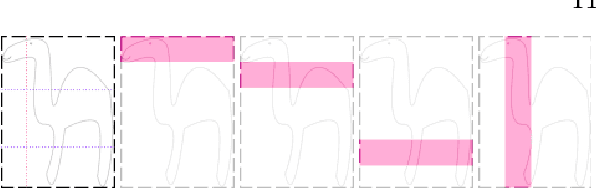
Abstract:Today, deep convolutional neural networks (CNNs) have demonstrated state of the art performance for supervised medical image segmentation, across various imaging modalities and tasks. Despite early success, segmentation networks may still generate anatomically aberrant segmentations, with holes or inaccuracies near the object boundaries. To mitigate this effect, recent research works have focused on incorporating spatial information or prior knowledge to enforce anatomically plausible segmentation. If the integration of prior knowledge in image segmentation is not a new topic in classical optimization approaches, it is today an increasing trend in CNN based image segmentation, as shown by the growing literature on the topic. In this survey, we focus on high level prior, embedded at the loss function level. We categorize the articles according to the nature of the prior: the object shape, size, topology, and the inter-regions constraints. We highlight strengths and limitations of current approaches, discuss the challenge related to the design and the integration of prior-based losses, and the optimization strategies, and draw future research directions.
Pattern Spotting in Historical Documents Using Convolutional Models
Jun 20, 2019



Abstract:Pattern spotting consists of searching in a collection of historical document images for occurrences of a graphical object using an image query. Contrary to object detection, no prior information nor predefined class is given about the query so training a model of the object is not feasible. In this paper, a convolutional neural network approach is proposed to tackle this problem. We use RetinaNet as a feature extractor to obtain multiscale embeddings of the regions of the documents and also for the queries. Experiments conducted on the DocExplore dataset show that our proposal is better at locating patterns and requires less storage for indexing images than the state-of-the-art system, but fails at retrieving some pages containing multiple instances of the query.
Medical Image Synthesis with Context-Aware Generative Adversarial Networks
Dec 16, 2016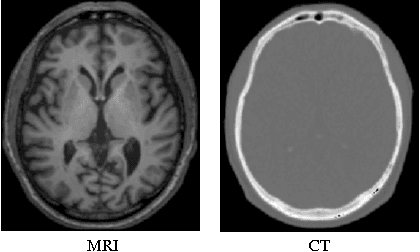
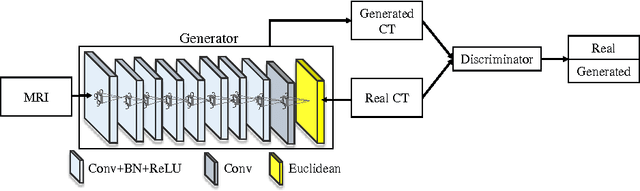
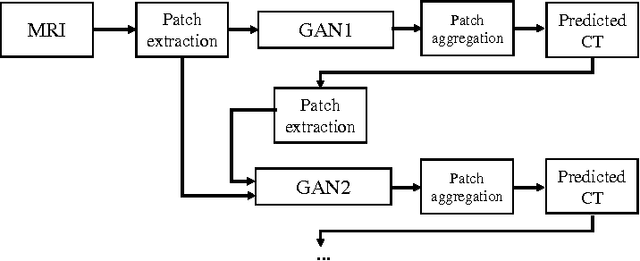
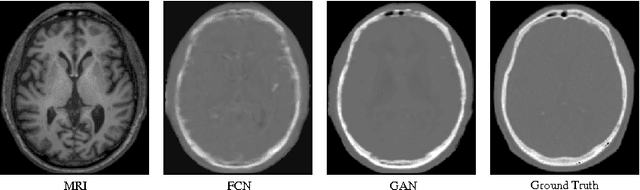
Abstract:Computed tomography (CT) is critical for various clinical applications, e.g., radiotherapy treatment planning and also PET attenuation correction. However, CT exposes radiation during acquisition, which may cause side effects to patients. Compared to CT, magnetic resonance imaging (MRI) is much safer and does not involve any radiations. Therefore, recently, researchers are greatly motivated to estimate CT image from its corresponding MR image of the same subject for the case of radiotherapy planning. In this paper, we propose a data-driven approach to address this challenging problem. Specifically, we train a fully convolutional network to generate CT given an MR image. To better model the nonlinear relationship from MRI to CT and to produce more realistic images, we propose to use the adversarial training strategy and an image gradient difference loss function. We further apply AutoContext Model to implement a context-aware generative adversarial network. Experimental results show that our method is accurate and robust for predicting CT images from MRI images, and also outperforms three state-of-the-art methods under comparison.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge