Rosana El Jurdi
SMILE-UHURA Challenge -- Small Vessel Segmentation at Mesoscopic Scale from Ultra-High Resolution 7T Magnetic Resonance Angiograms
Nov 14, 2024Abstract:The human brain receives nutrients and oxygen through an intricate network of blood vessels. Pathology affecting small vessels, at the mesoscopic scale, represents a critical vulnerability within the cerebral blood supply and can lead to severe conditions, such as Cerebral Small Vessel Diseases. The advent of 7 Tesla MRI systems has enabled the acquisition of higher spatial resolution images, making it possible to visualise such vessels in the brain. However, the lack of publicly available annotated datasets has impeded the development of robust, machine learning-driven segmentation algorithms. To address this, the SMILE-UHURA challenge was organised. This challenge, held in conjunction with the ISBI 2023, in Cartagena de Indias, Colombia, aimed to provide a platform for researchers working on related topics. The SMILE-UHURA challenge addresses the gap in publicly available annotated datasets by providing an annotated dataset of Time-of-Flight angiography acquired with 7T MRI. This dataset was created through a combination of automated pre-segmentation and extensive manual refinement. In this manuscript, sixteen submitted methods and two baseline methods are compared both quantitatively and qualitatively on two different datasets: held-out test MRAs from the same dataset as the training data (with labels kept secret) and a separate 7T ToF MRA dataset where both input volumes and labels are kept secret. The results demonstrate that most of the submitted deep learning methods, trained on the provided training dataset, achieved reliable segmentation performance. Dice scores reached up to 0.838 $\pm$ 0.066 and 0.716 $\pm$ 0.125 on the respective datasets, with an average performance of up to 0.804 $\pm$ 0.15.
Fourier Disentangled Multimodal Prior Knowledge Fusion for Red Nucleus Segmentation in Brain MRI
Nov 02, 2022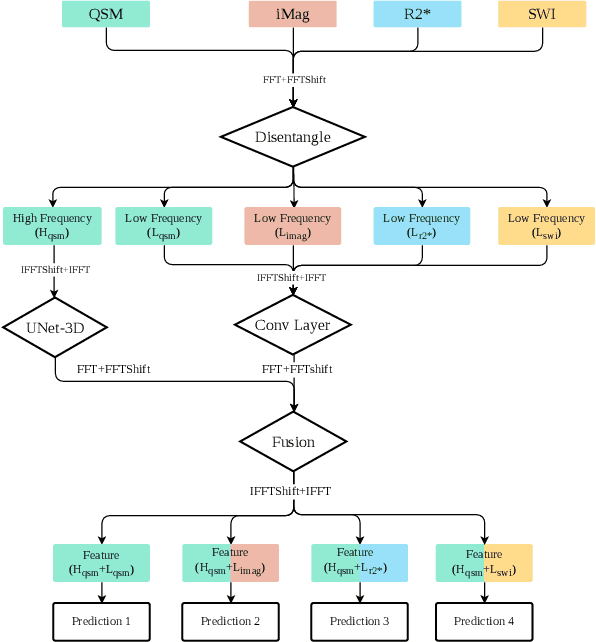
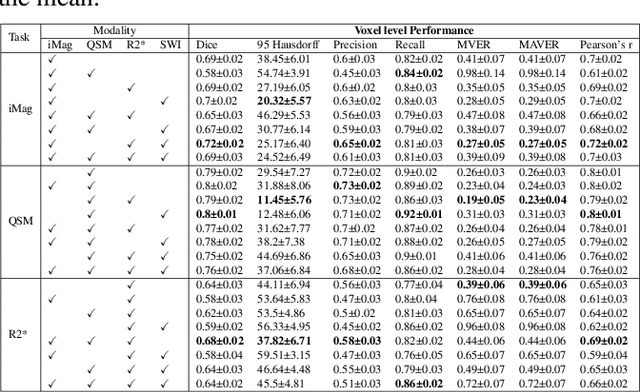
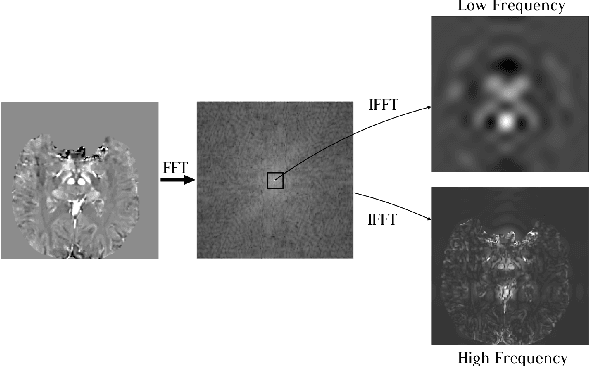
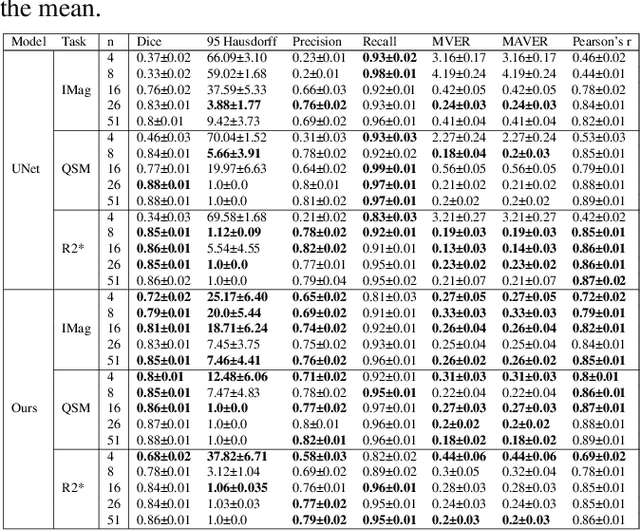
Abstract:Early and accurate diagnosis of parkinsonian syndromes is critical to provide appropriate care to patients and for inclusion in therapeutic trials. The red nucleus is a structure of the midbrain that plays an important role in these disorders. It can be visualized using iron-sensitive magnetic resonance imaging (MRI) sequences. Different iron-sensitive contrasts can be produced with MRI. Combining such multimodal data has the potential to improve segmentation of the red nucleus. Current multimodal segmentation algorithms are computationally consuming, cannot deal with missing modalities and need annotations for all modalities. In this paper, we propose a new model that integrates prior knowledge from different contrasts for red nucleus segmentation. The method consists of three main stages. First, it disentangles the image into high-level information representing the brain structure, and low-frequency information representing the contrast. The high-frequency information is then fed into a network to learn anatomical features, while the list of multimodal low-frequency information is processed by another module. Finally, feature fusion is performed to complete the segmentation task. The proposed method was used with several iron-sensitive contrasts (iMag, QSM, R2*, SWI). Experiments demonstrate that our proposed model substantially outperforms a baseline UNet model when the training set size is very small.
How precise are performance estimates for typical medical image segmentation tasks?
Nov 01, 2022Abstract:An important issue in medical image processing is to be able to estimate not only the performances of algorithms but also the precision of the estimation of these performances. Reporting precision typically amounts to reporting standard-error of the mean (SEM) or equivalently confidence intervals. However, this is rarely done in medical image segmentation studies. In this paper, we aim to estimate what is the typical confidence that can be expected in such studies. To that end, we first perform experiments for Dice metric estimation using a standard deep learning model (U-net) and a classical task from the Medical Segmentation Decathlon. We extensively study precision estimation using both Gaussian assumption and bootstrapping (which does not require any assumption on the distribution). We then perform simulations for other test set sizes and performance spreads. Overall, our work shows that small test sets lead to wide confidence intervals (e.g. $\sim$8 points of Dice for 20 samples with $\sigma \simeq 10$).
Effect of Prior-based Losses on Segmentation Performance: A Benchmark
Jan 12, 2022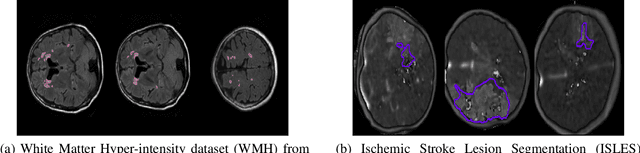
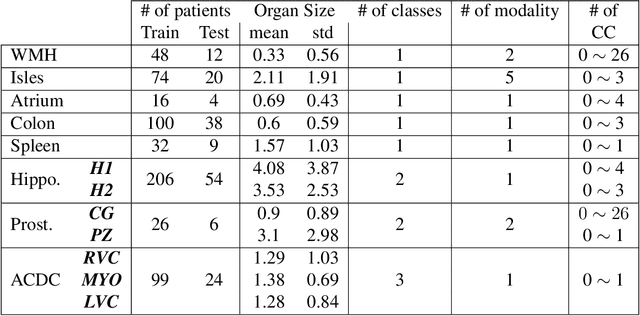
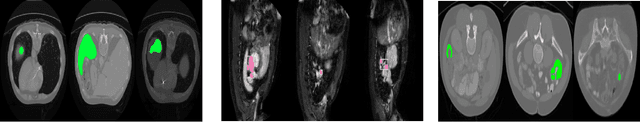

Abstract:Today, deep convolutional neural networks (CNNs) have demonstrated state-of-the-art performance for medical image segmentation, on various imaging modalities and tasks. Despite early success, segmentation networks may still generate anatomically aberrant segmentations, with holes or inaccuracies near the object boundaries. To enforce anatomical plausibility, recent research studies have focused on incorporating prior knowledge such as object shape or boundary, as constraints in the loss function. Prior integrated could be low-level referring to reformulated representations extracted from the ground-truth segmentations, or high-level representing external medical information such as the organ's shape or size. Over the past few years, prior-based losses exhibited a rising interest in the research field since they allow integration of expert knowledge while still being architecture-agnostic. However, given the diversity of prior-based losses on different medical imaging challenges and tasks, it has become hard to identify what loss works best for which dataset. In this paper, we establish a benchmark of recent prior-based losses for medical image segmentation. The main objective is to provide intuition onto which losses to choose given a particular task or dataset. To this end, four low-level and high-level prior-based losses are selected. The considered losses are validated on 8 different datasets from a variety of medical image segmentation challenges including the Decathlon, the ISLES and the WMH challenge. Results show that whereas low-level prior-based losses can guarantee an increase in performance over the Dice loss baseline regardless of the dataset characteristics, high-level prior-based losses can increase anatomical plausibility as per data characteristics.
High-level Prior-based Loss Functions for Medical Image Segmentation: A Survey
Nov 22, 2020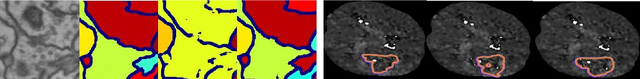
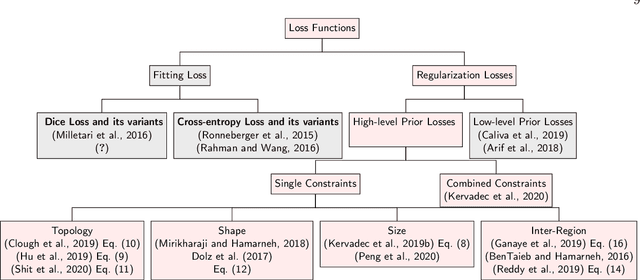
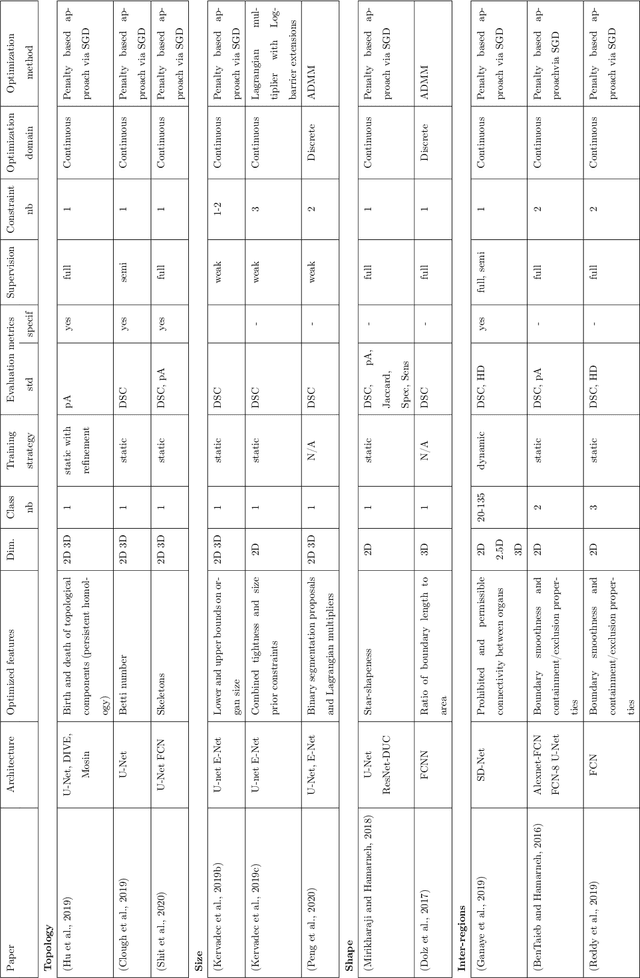
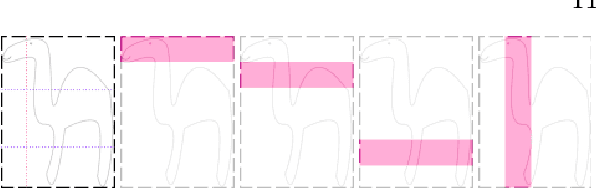
Abstract:Today, deep convolutional neural networks (CNNs) have demonstrated state of the art performance for supervised medical image segmentation, across various imaging modalities and tasks. Despite early success, segmentation networks may still generate anatomically aberrant segmentations, with holes or inaccuracies near the object boundaries. To mitigate this effect, recent research works have focused on incorporating spatial information or prior knowledge to enforce anatomically plausible segmentation. If the integration of prior knowledge in image segmentation is not a new topic in classical optimization approaches, it is today an increasing trend in CNN based image segmentation, as shown by the growing literature on the topic. In this survey, we focus on high level prior, embedded at the loss function level. We categorize the articles according to the nature of the prior: the object shape, size, topology, and the inter-regions constraints. We highlight strengths and limitations of current approaches, discuss the challenge related to the design and the integration of prior-based losses, and the optimization strategies, and draw future research directions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge