Romain Valabregue
SMILE-UHURA Challenge -- Small Vessel Segmentation at Mesoscopic Scale from Ultra-High Resolution 7T Magnetic Resonance Angiograms
Nov 14, 2024Abstract:The human brain receives nutrients and oxygen through an intricate network of blood vessels. Pathology affecting small vessels, at the mesoscopic scale, represents a critical vulnerability within the cerebral blood supply and can lead to severe conditions, such as Cerebral Small Vessel Diseases. The advent of 7 Tesla MRI systems has enabled the acquisition of higher spatial resolution images, making it possible to visualise such vessels in the brain. However, the lack of publicly available annotated datasets has impeded the development of robust, machine learning-driven segmentation algorithms. To address this, the SMILE-UHURA challenge was organised. This challenge, held in conjunction with the ISBI 2023, in Cartagena de Indias, Colombia, aimed to provide a platform for researchers working on related topics. The SMILE-UHURA challenge addresses the gap in publicly available annotated datasets by providing an annotated dataset of Time-of-Flight angiography acquired with 7T MRI. This dataset was created through a combination of automated pre-segmentation and extensive manual refinement. In this manuscript, sixteen submitted methods and two baseline methods are compared both quantitatively and qualitatively on two different datasets: held-out test MRAs from the same dataset as the training data (with labels kept secret) and a separate 7T ToF MRA dataset where both input volumes and labels are kept secret. The results demonstrate that most of the submitted deep learning methods, trained on the provided training dataset, achieved reliable segmentation performance. Dice scores reached up to 0.838 $\pm$ 0.066 and 0.716 $\pm$ 0.125 on the respective datasets, with an average performance of up to 0.804 $\pm$ 0.15.
PyCellMech: A shape-based feature extraction pipeline for use in medical and biological studies
May 24, 2024Abstract:Summary: Medical researchers obtain knowledge about the prevention and treatment of disability and disease using physical measurements and image data. To assist in this endeavor, feature extraction packages are available that are designed to collect data from the image structure. In this study, we aim to augment current works by adding to the current mix of shape-based features. The significance of shape-based features has been explored extensively in research for several decades, but there is no single package available in which all shape-related features can be extracted easily by the researcher. PyCellMech has been crafted to address this gap. The PyCellMech package extracts three classes of shape features, which are classified as one-dimensional, geometric, and polygonal. Future iterations will be expanded to include other feature classes, such as scale-space. Availability and implementation: PyCellMech is freely available at https://github.com/icm-dac/pycellmech.
Multi-Center Fetal Brain Tissue Annotation (FeTA) Challenge 2022 Results
Feb 08, 2024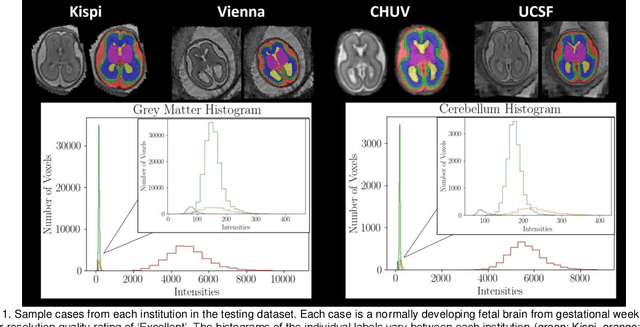
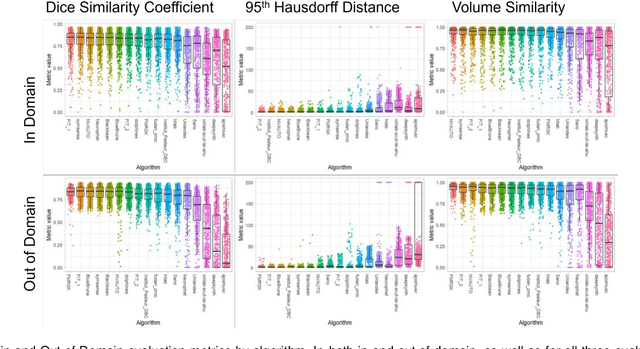
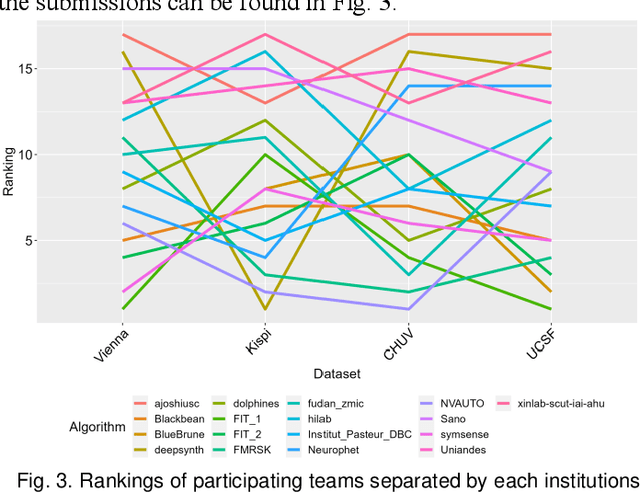
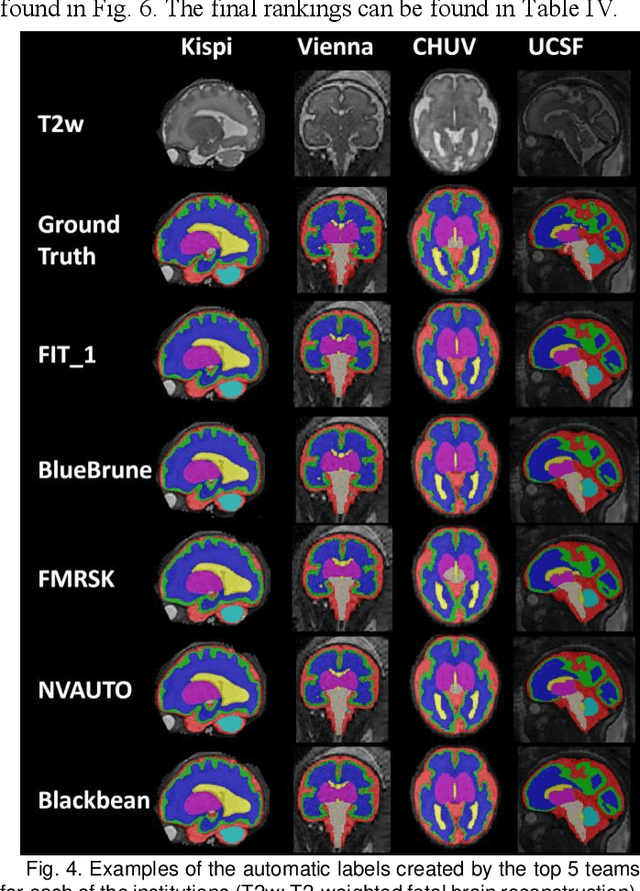
Abstract:Segmentation is a critical step in analyzing the developing human fetal brain. There have been vast improvements in automatic segmentation methods in the past several years, and the Fetal Brain Tissue Annotation (FeTA) Challenge 2021 helped to establish an excellent standard of fetal brain segmentation. However, FeTA 2021 was a single center study, and the generalizability of algorithms across different imaging centers remains unsolved, limiting real-world clinical applicability. The multi-center FeTA Challenge 2022 focuses on advancing the generalizability of fetal brain segmentation algorithms for magnetic resonance imaging (MRI). In FeTA 2022, the training dataset contained images and corresponding manually annotated multi-class labels from two imaging centers, and the testing data contained images from these two imaging centers as well as two additional unseen centers. The data from different centers varied in many aspects, including scanners used, imaging parameters, and fetal brain super-resolution algorithms applied. 16 teams participated in the challenge, and 17 algorithms were evaluated. Here, a detailed overview and analysis of the challenge results are provided, focusing on the generalizability of the submissions. Both in- and out of domain, the white matter and ventricles were segmented with the highest accuracy, while the most challenging structure remains the cerebral cortex due to anatomical complexity. The FeTA Challenge 2022 was able to successfully evaluate and advance generalizability of multi-class fetal brain tissue segmentation algorithms for MRI and it continues to benchmark new algorithms. The resulting new methods contribute to improving the analysis of brain development in utero.
Frequency Disentangled Learning for Segmentation of Midbrain Structures from Quantitative Susceptibility Mapping Data
Feb 25, 2023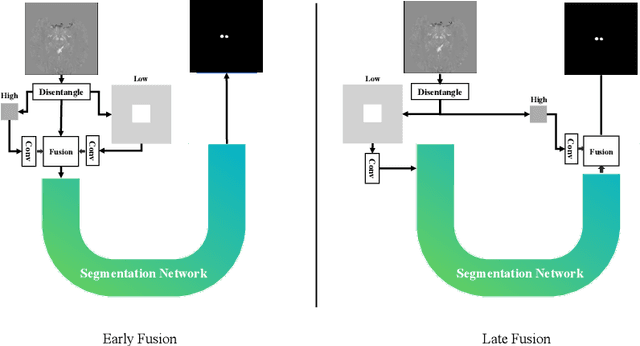

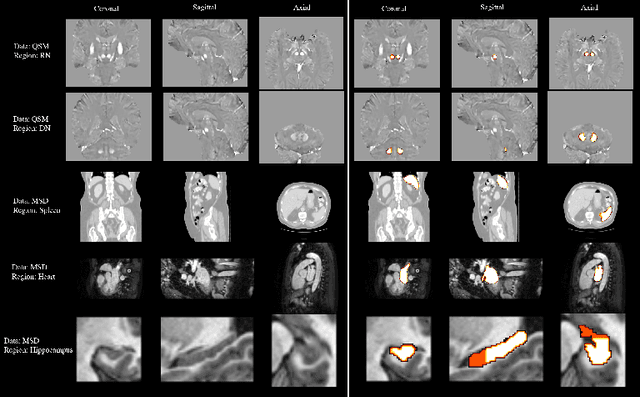
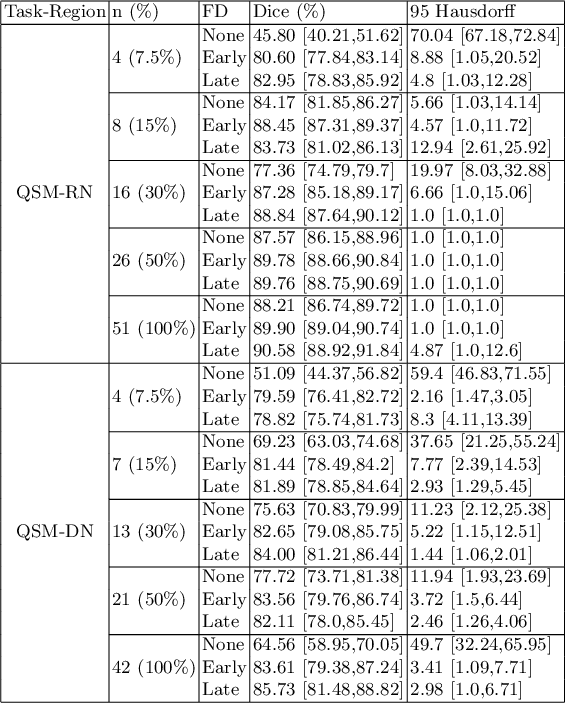
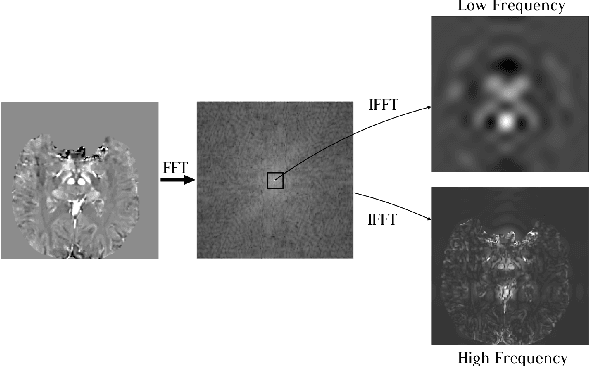
Abstract:One often lacks sufficient annotated samples for training deep segmentation models. This is in particular the case for less common imaging modalities such as Quantitative Susceptibility Mapping (QSM). It has been shown that deep models tend to fit the target function from low to high frequencies. One may hypothesize that such property can be leveraged for better training of deep learning models. In this paper, we exploit this property to propose a new training method based on frequency-domain disentanglement. It consists of two main steps: i) disentangling the image into high- and low-frequency parts and feature learning; ii) frequency-domain fusion to complete the task. The approach can be used with any backbone segmentation network. We apply the approach to the segmentation of the red and dentate nuclei from QSM data which is particularly relevant for the study of parkinsonian syndromes. We demonstrate that the proposed method provides considerable performance improvements for these tasks. We further applied it to three public datasets from the Medical Segmentation Decathlon (MSD) challenge. For two MSD tasks, it provided smaller but still substantial improvements (up to 7 points of Dice), especially under small training set situations.
Fourier Disentangled Multimodal Prior Knowledge Fusion for Red Nucleus Segmentation in Brain MRI
Nov 02, 2022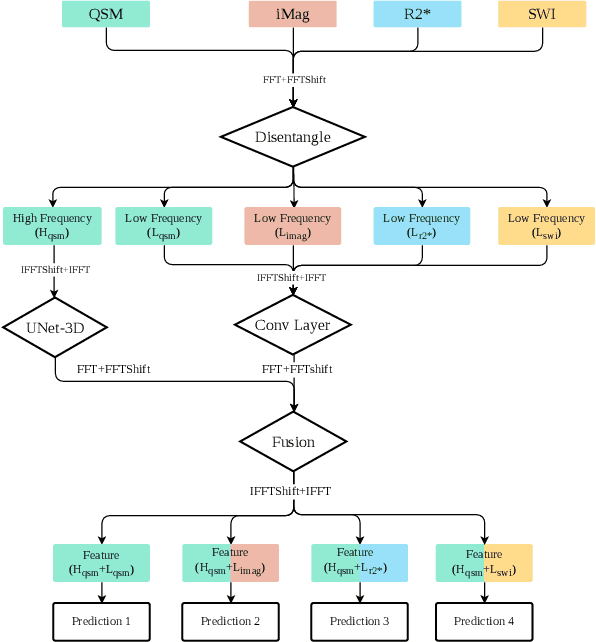
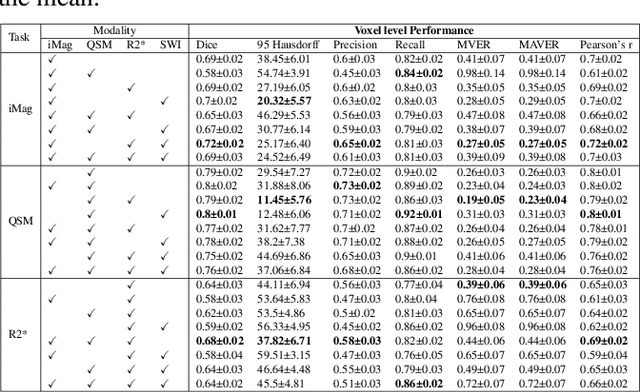
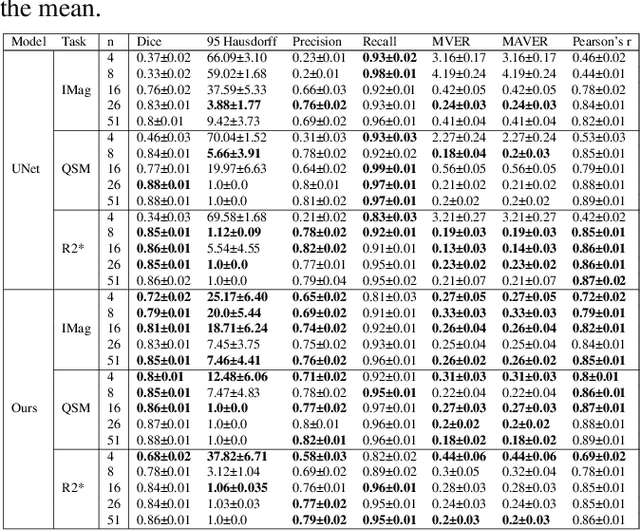
Abstract:Early and accurate diagnosis of parkinsonian syndromes is critical to provide appropriate care to patients and for inclusion in therapeutic trials. The red nucleus is a structure of the midbrain that plays an important role in these disorders. It can be visualized using iron-sensitive magnetic resonance imaging (MRI) sequences. Different iron-sensitive contrasts can be produced with MRI. Combining such multimodal data has the potential to improve segmentation of the red nucleus. Current multimodal segmentation algorithms are computationally consuming, cannot deal with missing modalities and need annotations for all modalities. In this paper, we propose a new model that integrates prior knowledge from different contrasts for red nucleus segmentation. The method consists of three main stages. First, it disentangles the image into high-level information representing the brain structure, and low-frequency information representing the contrast. The high-frequency information is then fed into a network to learn anatomical features, while the list of multimodal low-frequency information is processed by another module. Finally, feature fusion is performed to complete the segmentation task. The proposed method was used with several iron-sensitive contrasts (iMag, QSM, R2*, SWI). Experiments demonstrate that our proposed model substantially outperforms a baseline UNet model when the training set size is very small.
Learning brain MRI quality control: a multi-factorial generalization problem
May 31, 2022
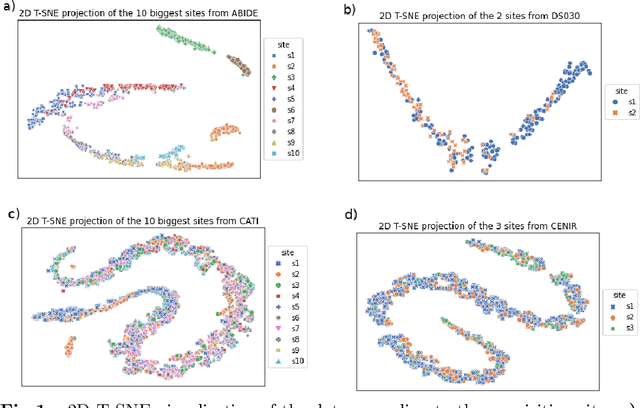
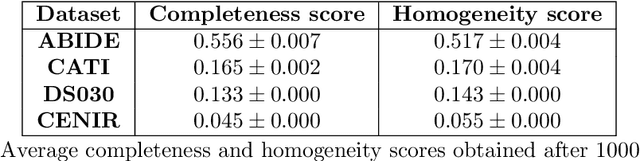
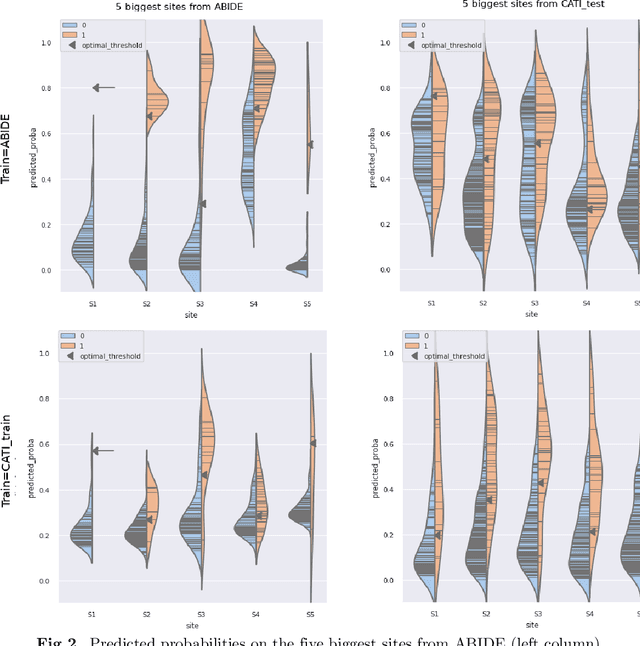
Abstract:Due to the growing number of MRI data, automated quality control (QC) has become essential, especially for larger scale analysis. Several attempts have been made in order to develop reliable and scalable QC pipelines. However, the generalization of these methods on new data independent of those used for learning is a difficult problem because of the biases inherent in MRI data. This work aimed at evaluating the performances of the MRIQC pipeline on various large-scale datasets (ABIDE, N = 1102 and CATI derived datasets, N = 9037) used for both training and evaluation purposes. We focused our analysis on the MRIQC preprocessing steps and tested the pipeline with and without them. We further analyzed the site-wise and study-wise predicted classification probability distributions of the models without preprocessing trained on ABIDE and CATI data. Our main results were that a model using features extracted from MRIQC without preprocessing yielded the best results when trained and evaluated on large multi-center datasets with a heterogeneous population (an improvement of the ROC-AUC score on unseen data of 0.10 for the model trained on a subset of the CATI dataset). We concluded that a model trained with data from a heterogeneous population, such as the CATI dataset, provides the best scores on unseen data. In spite of the performance improvement, the generalization abilities of the models remain questionable when looking at the site-wise/study-wise probability predictions and the optimal classification threshold derived from them.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge