Stéphane Lehéricy
CENIR, ICM
Frequency Disentangled Learning for Segmentation of Midbrain Structures from Quantitative Susceptibility Mapping Data
Feb 25, 2023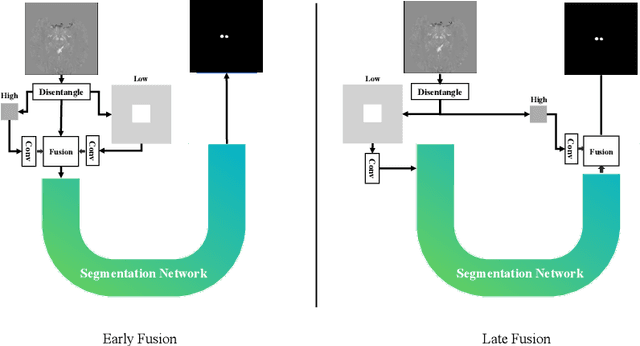

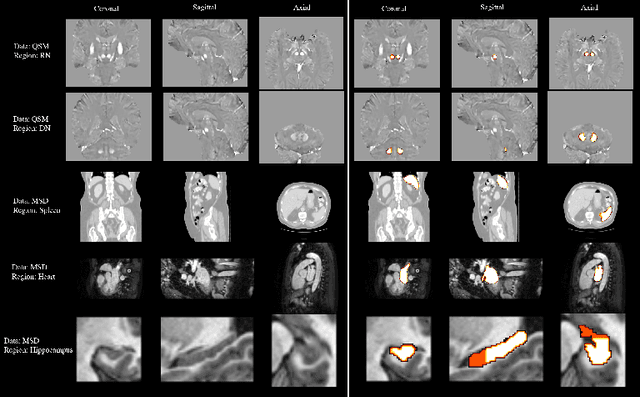
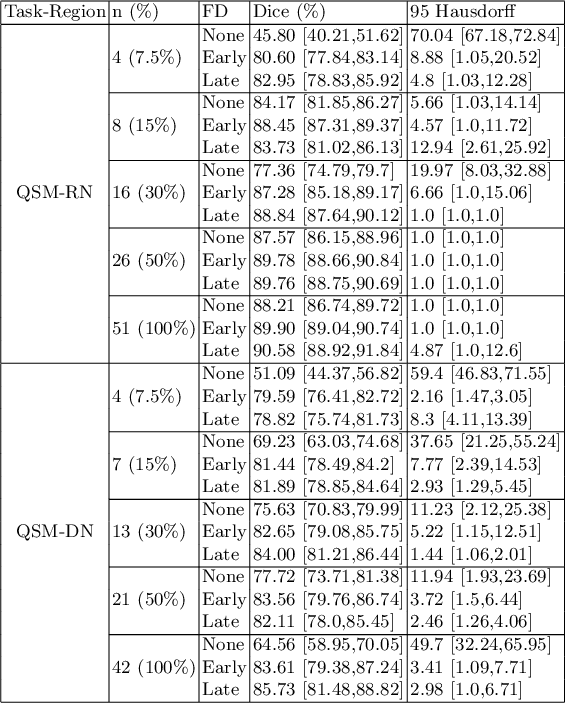
Abstract:One often lacks sufficient annotated samples for training deep segmentation models. This is in particular the case for less common imaging modalities such as Quantitative Susceptibility Mapping (QSM). It has been shown that deep models tend to fit the target function from low to high frequencies. One may hypothesize that such property can be leveraged for better training of deep learning models. In this paper, we exploit this property to propose a new training method based on frequency-domain disentanglement. It consists of two main steps: i) disentangling the image into high- and low-frequency parts and feature learning; ii) frequency-domain fusion to complete the task. The approach can be used with any backbone segmentation network. We apply the approach to the segmentation of the red and dentate nuclei from QSM data which is particularly relevant for the study of parkinsonian syndromes. We demonstrate that the proposed method provides considerable performance improvements for these tasks. We further applied it to three public datasets from the Medical Segmentation Decathlon (MSD) challenge. For two MSD tasks, it provided smaller but still substantial improvements (up to 7 points of Dice), especially under small training set situations.
Fourier Disentangled Multimodal Prior Knowledge Fusion for Red Nucleus Segmentation in Brain MRI
Nov 02, 2022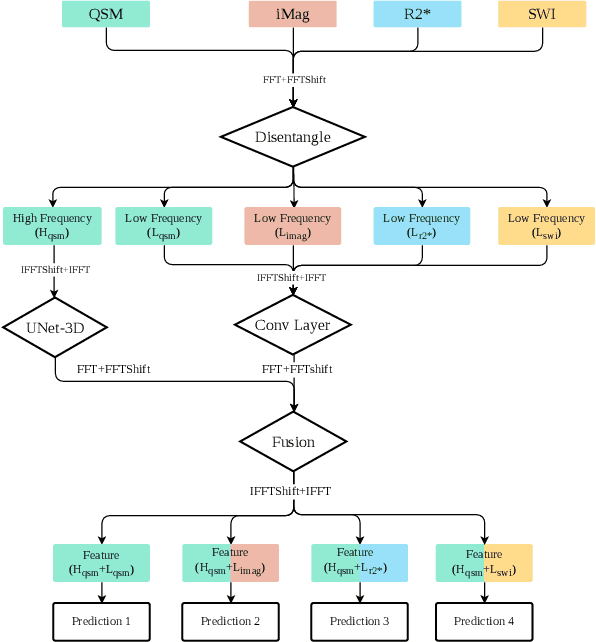
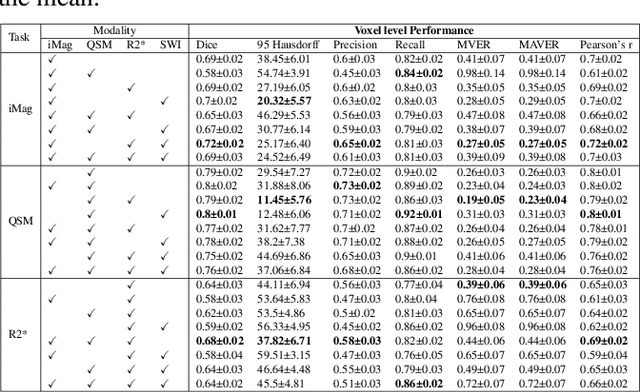
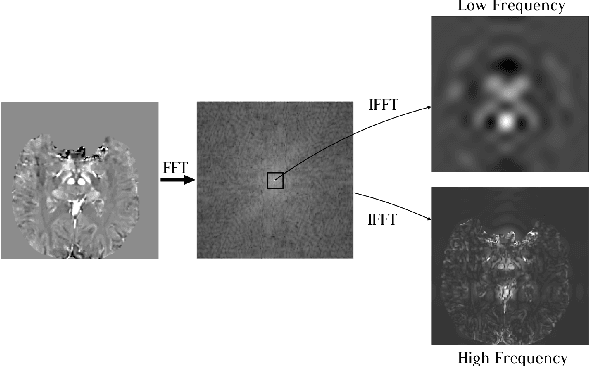
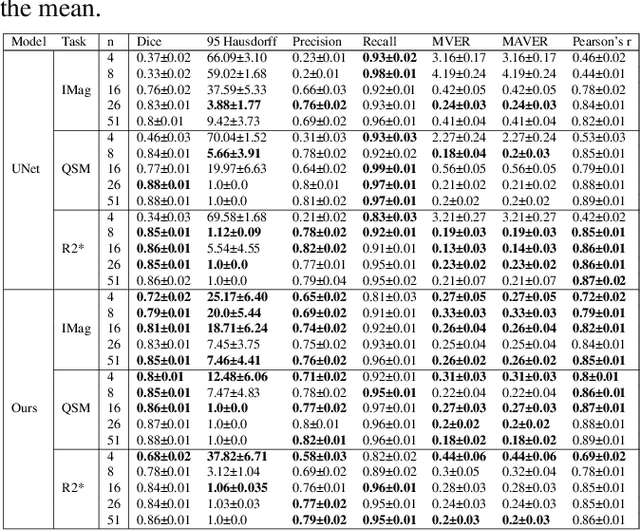
Abstract:Early and accurate diagnosis of parkinsonian syndromes is critical to provide appropriate care to patients and for inclusion in therapeutic trials. The red nucleus is a structure of the midbrain that plays an important role in these disorders. It can be visualized using iron-sensitive magnetic resonance imaging (MRI) sequences. Different iron-sensitive contrasts can be produced with MRI. Combining such multimodal data has the potential to improve segmentation of the red nucleus. Current multimodal segmentation algorithms are computationally consuming, cannot deal with missing modalities and need annotations for all modalities. In this paper, we propose a new model that integrates prior knowledge from different contrasts for red nucleus segmentation. The method consists of three main stages. First, it disentangles the image into high-level information representing the brain structure, and low-frequency information representing the contrast. The high-frequency information is then fed into a network to learn anatomical features, while the list of multimodal low-frequency information is processed by another module. Finally, feature fusion is performed to complete the segmentation task. The proposed method was used with several iron-sensitive contrasts (iMag, QSM, R2*, SWI). Experiments demonstrate that our proposed model substantially outperforms a baseline UNet model when the training set size is very small.
Learning brain MRI quality control: a multi-factorial generalization problem
May 31, 2022
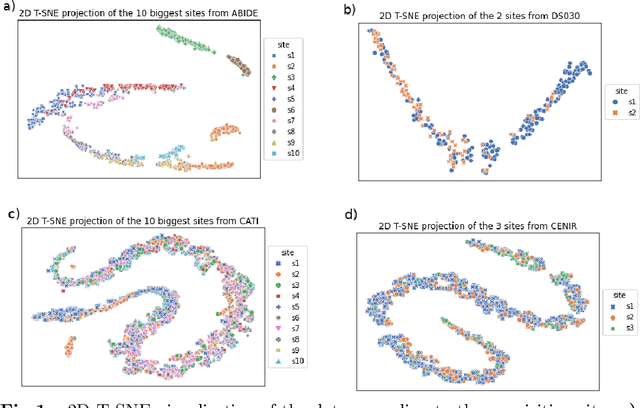
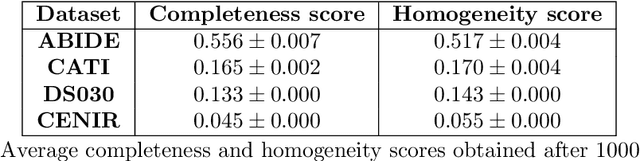
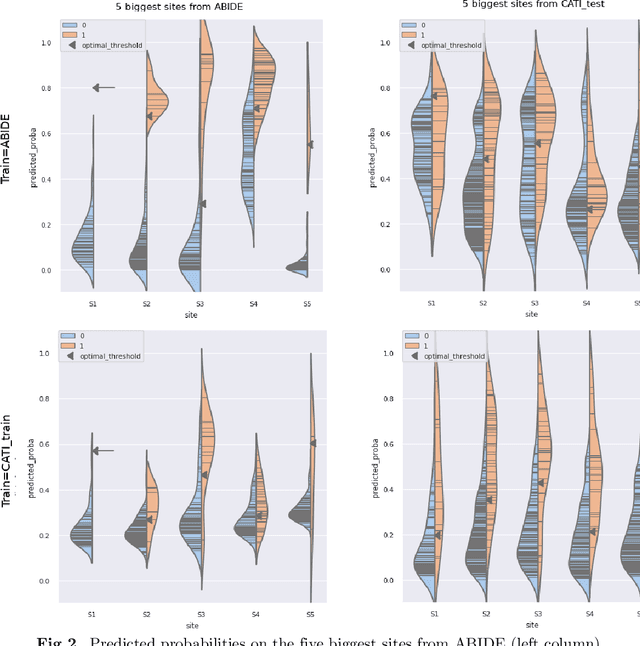
Abstract:Due to the growing number of MRI data, automated quality control (QC) has become essential, especially for larger scale analysis. Several attempts have been made in order to develop reliable and scalable QC pipelines. However, the generalization of these methods on new data independent of those used for learning is a difficult problem because of the biases inherent in MRI data. This work aimed at evaluating the performances of the MRIQC pipeline on various large-scale datasets (ABIDE, N = 1102 and CATI derived datasets, N = 9037) used for both training and evaluation purposes. We focused our analysis on the MRIQC preprocessing steps and tested the pipeline with and without them. We further analyzed the site-wise and study-wise predicted classification probability distributions of the models without preprocessing trained on ABIDE and CATI data. Our main results were that a model using features extracted from MRIQC without preprocessing yielded the best results when trained and evaluated on large multi-center datasets with a heterogeneous population (an improvement of the ROC-AUC score on unseen data of 0.10 for the model trained on a subset of the CATI dataset). We concluded that a model trained with data from a heterogeneous population, such as the CATI dataset, provides the best scores on unseen data. In spite of the performance improvement, the generalization abilities of the models remain questionable when looking at the site-wise/study-wise probability predictions and the optimal classification threshold derived from them.
Multidimensional classification of hippocampal shape features discriminates Alzheimer's disease and mild cognitive impairment from normal aging
Jul 19, 2017



Abstract:We describe a new method to automatically discriminate between patients with Alzheimer's disease (AD) or mild cognitive impairment (MCI) and elderly controls, based on multidimensional classification of hippocampal shape features. This approach uses spherical harmonics (SPHARM) coefficients to model the shape of the hippocampi, which are segmented from magnetic resonance images (MRI) using a fully automatic method that we previously developed. SPHARM coefficients are used as features in a classification procedure based on support vector machines (SVM). The most relevant features for classification are selected using a bagging strategy. We evaluate the accuracy of our method in a group of 23 patients with AD (10 males, 13 females, age $\pm$ standard-deviation (SD) = 73 $\pm$ 6 years, mini-mental score (MMS) = 24.4 $\pm$ 2.8), 23 patients with amnestic MCI (10 males, 13 females, age $\pm$ SD = 74 $\pm$ 8 years, MMS = 27.3 $\pm$ 1.4) and 25 elderly healthy controls (13 males, 12 females, age $\pm$ SD = 64 $\pm$ 8 years), using leave-one-out cross-validation. For AD vs controls, we obtain a correct classification rate of 94%, a sensitivity of 96%, and a specificity of 92%. For MCI vs controls, we obtain a classification rate of 83%, a sensitivity of 83%, and a specificity of 84%. This accuracy is superior to that of hippocampal volumetry and is comparable to recently published SVM-based whole-brain classification methods, which relied on a different strategy. This new method may become a useful tool to assist in the diagnosis of Alzheimer's disease.
* Data used in the preparation of this article were obtained from the Alzheimer's Disease Neuroimaging Initiative (ADNI) database
Robust imaging of hippocampal inner structure at 7T: in vivo acquisition protocol and methodological choices
May 09, 2016



Abstract:OBJECTIVE:Motion-robust multi-slab imaging of hippocampal inner structure in vivo at 7T.MATERIALS AND METHODS:Motion is a crucial issue for ultra-high resolution imaging, such as can be achieved with 7T MRI. An acquisition protocol was designed for imaging hippocampal inner structure at 7T. It relies on a compromise between anatomical details visibility and robustness to motion. In order to reduce acquisition time and motion artifacts, the full slab covering the hippocampus was split into separate slabs with lower acquisition time. A robust registration approach was implemented to combine the acquired slabs within a final 3D-consistent high-resolution slab covering the whole hippocampus. Evaluation was performed on 50 subjects overall, made of three groups of subjects acquired using three acquisition settings; it focused on three issues: visibility of hippocampal inner structure, robustness to motion artifacts and registration procedure performance.RESULTS:Overall, T2-weighted acquisitions with interleaved slabs proved robust. Multi-slab registration yielded high quality datasets in 96 % of the subjects, thus compatible with further analyses of hippocampal inner structure.CONCLUSION:Multi-slab acquisition and registration setting is efficient for reducing acquisition time and consequently motion artifacts for ultra-high resolution imaging of the inner structure of the hippocampus.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge