Learning brain MRI quality control: a multi-factorial generalization problem
Paper and Code
May 31, 2022
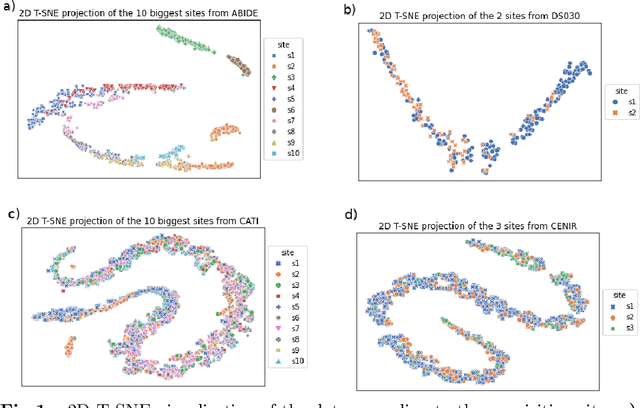
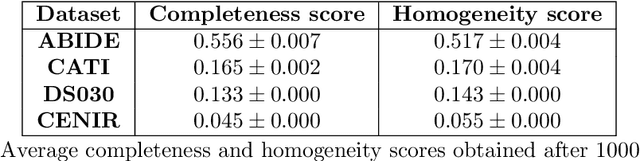
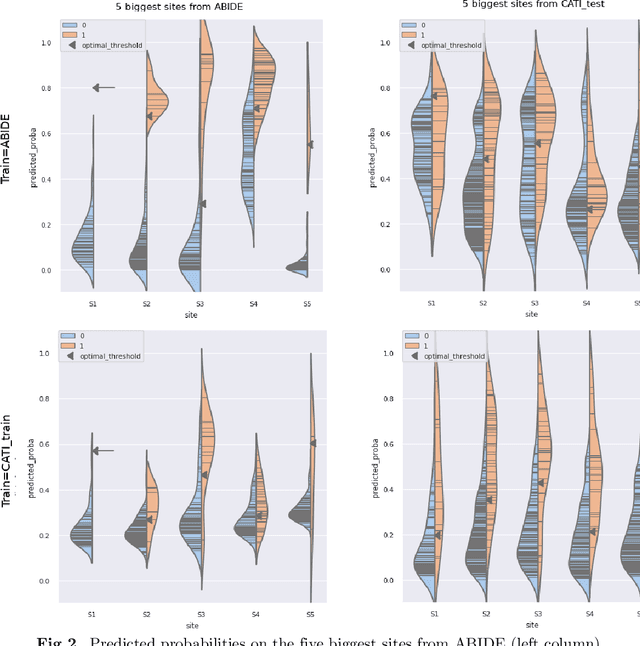
Due to the growing number of MRI data, automated quality control (QC) has become essential, especially for larger scale analysis. Several attempts have been made in order to develop reliable and scalable QC pipelines. However, the generalization of these methods on new data independent of those used for learning is a difficult problem because of the biases inherent in MRI data. This work aimed at evaluating the performances of the MRIQC pipeline on various large-scale datasets (ABIDE, N = 1102 and CATI derived datasets, N = 9037) used for both training and evaluation purposes. We focused our analysis on the MRIQC preprocessing steps and tested the pipeline with and without them. We further analyzed the site-wise and study-wise predicted classification probability distributions of the models without preprocessing trained on ABIDE and CATI data. Our main results were that a model using features extracted from MRIQC without preprocessing yielded the best results when trained and evaluated on large multi-center datasets with a heterogeneous population (an improvement of the ROC-AUC score on unseen data of 0.10 for the model trained on a subset of the CATI dataset). We concluded that a model trained with data from a heterogeneous population, such as the CATI dataset, provides the best scores on unseen data. In spite of the performance improvement, the generalization abilities of the models remain questionable when looking at the site-wise/study-wise probability predictions and the optimal classification threshold derived from them.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge