Marc Niethammer
On The Robustness of Foundational 3D Medical Image Segmentation Models Against Imprecise Visual Prompts
Jan 23, 2026Abstract:While 3D foundational models have shown promise for promptable segmentation of medical volumes, their robustness to imprecise prompts remains under-explored. In this work, we aim to address this gap by systematically studying the effect of various controlled perturbations of dense visual prompts, that closely mimic real-world imprecision. By conducting experiments with two recent foundational models on a multi-organ abdominal segmentation task, we reveal several facets of promptable medical segmentation, especially pertaining to reliance on visual shape and spatial cues, and the extent of resilience of models towards certain perturbations. Codes are available at: https://github.com/ucsdbiag/Prompt-Robustness-MedSegFMs
NOCTA: Non-Greedy Objective Cost-Tradeoff Acquisition for Longitudinal Data
Jul 16, 2025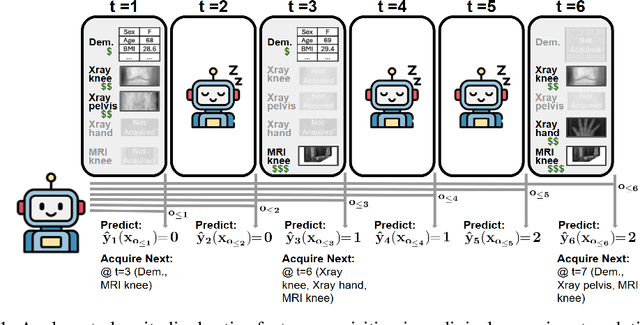
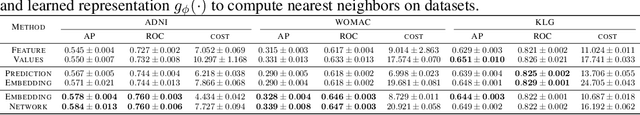
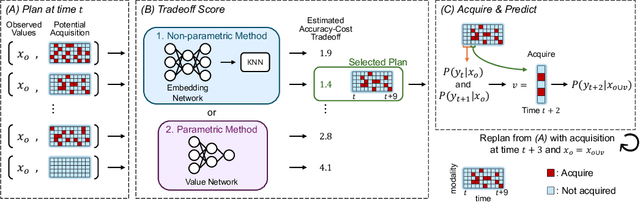
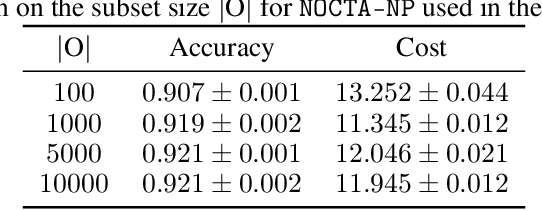
Abstract:In many critical applications, resource constraints limit the amount of information that can be gathered to make predictions. For example, in healthcare, patient data often spans diverse features ranging from lab tests to imaging studies. Each feature may carry different information and must be acquired at a respective cost of time, money, or risk to the patient. Moreover, temporal prediction tasks, where both instance features and labels evolve over time, introduce additional complexity in deciding when or what information is important. In this work, we propose NOCTA, a Non-Greedy Objective Cost-Tradeoff Acquisition method that sequentially acquires the most informative features at inference time while accounting for both temporal dynamics and acquisition cost. We first introduce a cohesive estimation target for our NOCTA setting, and then develop two complementary estimators: 1) a non-parametric method based on nearest neighbors to guide the acquisition (NOCTA-NP), and 2) a parametric method that directly predicts the utility of potential acquisitions (NOCTA-P). Experiments on synthetic and real-world medical datasets demonstrate that both NOCTA variants outperform existing baselines.
PPS-Ctrl: Controllable Sim-to-Real Translation for Colonoscopy Depth Estimation
Apr 23, 2025Abstract:Accurate depth estimation enhances endoscopy navigation and diagnostics, but obtaining ground-truth depth in clinical settings is challenging. Synthetic datasets are often used for training, yet the domain gap limits generalization to real data. We propose a novel image-to-image translation framework that preserves structure while generating realistic textures from clinical data. Our key innovation integrates Stable Diffusion with ControlNet, conditioned on a latent representation extracted from a Per-Pixel Shading (PPS) map. PPS captures surface lighting effects, providing a stronger structural constraint than depth maps. Experiments show our approach produces more realistic translations and improves depth estimation over GAN-based MI-CycleGAN. Our code is publicly accessible at https://github.com/anaxqx/PPS-Ctrl.
Zero-shot Domain Generalization of Foundational Models for 3D Medical Image Segmentation: An Experimental Study
Mar 28, 2025Abstract:Domain shift, caused by variations in imaging modalities and acquisition protocols, limits model generalization in medical image segmentation. While foundation models (FMs) trained on diverse large-scale data hold promise for zero-shot generalization, their application to volumetric medical data remains underexplored. In this study, we examine their ability towards domain generalization (DG), by conducting a comprehensive experimental study encompassing 6 medical segmentation FMs and 12 public datasets spanning multiple modalities and anatomies. Our findings reveal the potential of promptable FMs in bridging the domain gap via smart prompting techniques. Additionally, by probing into multiple facets of zero-shot DG, we offer valuable insights into the viability of FMs for DG and identify promising avenues for future research.
Downstream Analysis of Foundational Medical Vision Models for Disease Progression
Mar 21, 2025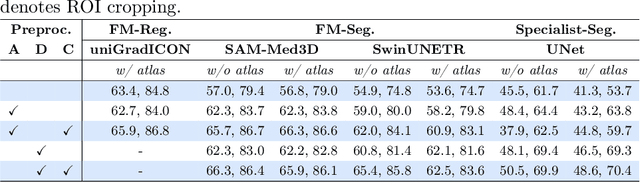
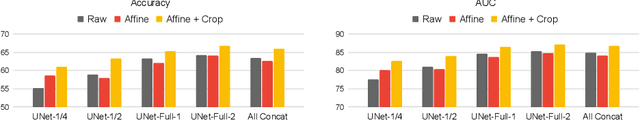
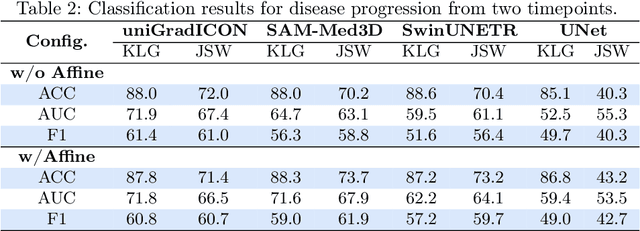
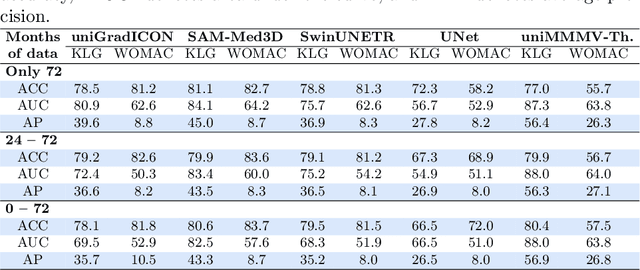
Abstract:Medical vision foundational models are used for a wide variety of tasks, including medical image segmentation and registration. This work evaluates the ability of these models to predict disease progression using a simple linear probe. We hypothesize that intermediate layer features of segmentation models capture structural information, while those of registration models encode knowledge of change over time. Beyond demonstrating that these features are useful for disease progression prediction, we also show that registration model features do not require spatially aligned input images. However, for segmentation models, spatial alignment is essential for optimal performance. Our findings highlight the importance of spatial alignment and the utility of foundation model features for image registration.
$\texttt{LucidAtlas}$: Learning Uncertainty-Aware, Covariate-Disentangled, Individualized Atlas Representations
Feb 12, 2025Abstract:The goal of this work is to develop principled techniques to extract information from high dimensional data sets with complex dependencies in areas such as medicine that can provide insight into individual as well as population level variation. We develop $\texttt{LucidAtlas}$, an approach that can represent spatially varying information, and can capture the influence of covariates as well as population uncertainty. As a versatile atlas representation, $\texttt{LucidAtlas}$ offers robust capabilities for covariate interpretation, individualized prediction, population trend analysis, and uncertainty estimation, with the flexibility to incorporate prior knowledge. Additionally, we discuss the trustworthiness and potential risks of neural additive models for analyzing dependent covariates and then introduce a marginalization approach to explain the dependence of an individual predictor on the models' response (the atlas). To validate our method, we demonstrate its generalizability on two medical datasets. Our findings underscore the critical role of by-construction interpretable models in advancing scientific discovery. Our code will be publicly available upon acceptance.
NFL-BA: Improving Endoscopic SLAM with Near-Field Light Bundle Adjustment
Dec 17, 2024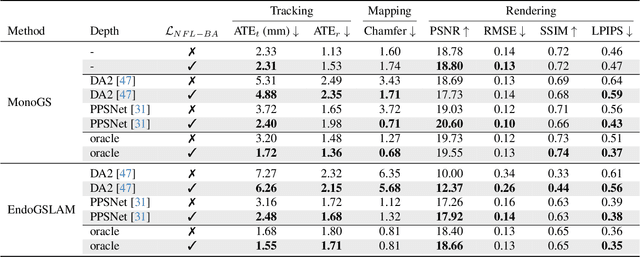
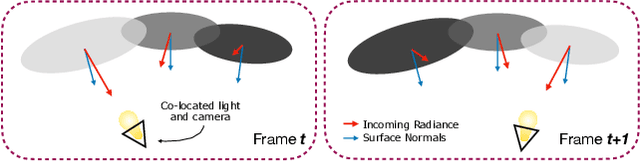
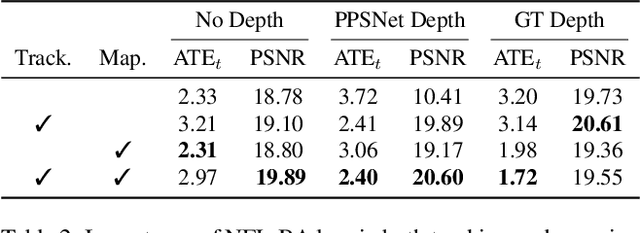
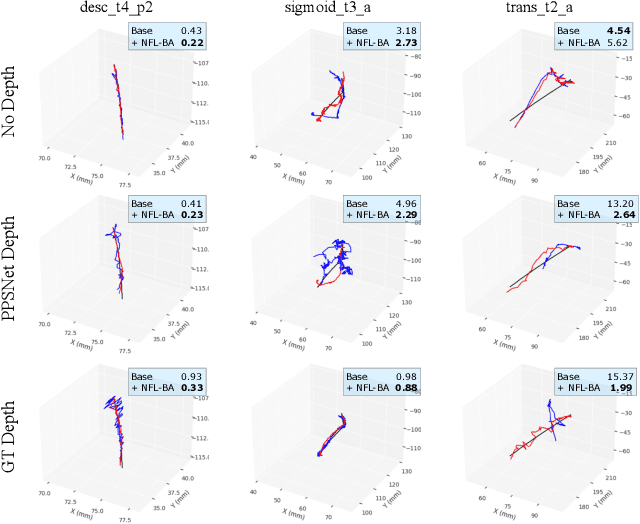
Abstract:Simultaneous Localization And Mapping (SLAM) from a monocular endoscopy video can enable autonomous navigation, guidance to unsurveyed regions, and 3D visualizations, which can significantly improve endoscopy experience for surgeons and patient outcomes. Existing dense SLAM algorithms often assume distant and static lighting and textured surfaces, and alternate between optimizing scene geometry and camera parameters by minimizing a photometric rendering loss, often called Photometric Bundle Adjustment. However, endoscopic environments exhibit dynamic near-field lighting due to the co-located light and camera moving extremely close to the surface, textureless surfaces, and strong specular reflections due to mucus layers. When not considered, these near-field lighting effects can cause significant performance reductions for existing SLAM algorithms from indoor/outdoor scenes when applied to endoscopy videos. To mitigate this problem, we introduce a new Near-Field Lighting Bundle Adjustment Loss $(L_{NFL-BA})$ that can also be alternatingly optimized, along with the Photometric Bundle Adjustment loss, such that the captured images' intensity variations match the relative distance and orientation between the surface and the co-located light and camera. We derive a general NFL-BA loss function for 3D Gaussian surface representations and demonstrate that adding $L_{NFL-BA}$ can significantly improve the tracking and mapping performance of two state-of-the-art 3DGS-SLAM systems, MonoGS (35% improvement in tracking, 48% improvement in mapping with predicted depth maps) and EndoGSLAM (22% improvement in tracking, marginal improvement in mapping with predicted depths), on the C3VD endoscopy dataset for colons. The project page is available at https://asdunnbe.github.io/NFL-BA/
LiVOS: Light Video Object Segmentation with Gated Linear Matching
Nov 05, 2024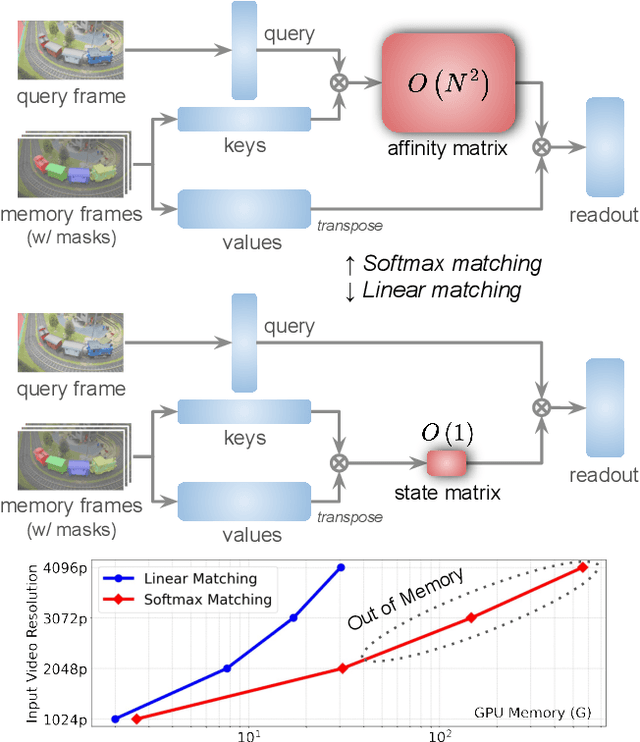
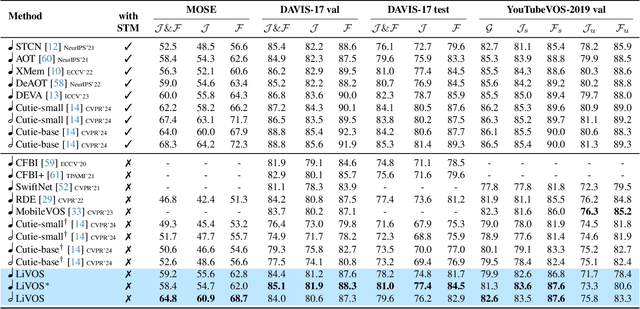
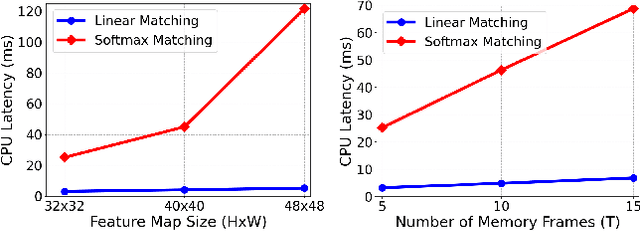
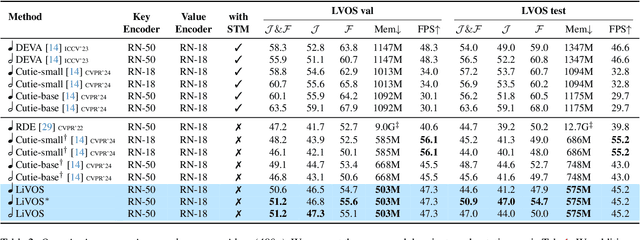
Abstract:Semi-supervised video object segmentation (VOS) has been largely driven by space-time memory (STM) networks, which store past frame features in a spatiotemporal memory to segment the current frame via softmax attention. However, STM networks face memory limitations due to the quadratic complexity of softmax matching, restricting their applicability as video length and resolution increase. To address this, we propose LiVOS, a lightweight memory network that employs linear matching via linear attention, reformulating memory matching into a recurrent process that reduces the quadratic attention matrix to a constant-size, spatiotemporal-agnostic 2D state. To enhance selectivity, we introduce gated linear matching, where a data-dependent gate matrix is multiplied with the state matrix to control what information to retain or discard. Experiments on diverse benchmarks demonstrated the effectiveness of our method. It achieved 64.8 J&F on MOSE and 85.1 J&F on DAVIS, surpassing all non-STM methods and narrowing the gap with STM-based approaches. For longer and higher-resolution videos, it matched STM-based methods with 53% less GPU memory and supports 4096p inference on a 32G consumer-grade GPU--a previously cost-prohibitive capability--opening the door for long and high-resolution video foundation models.
multiGradICON: A Foundation Model for Multimodal Medical Image Registration
Aug 01, 2024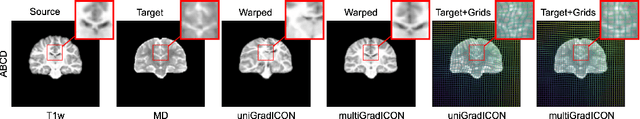
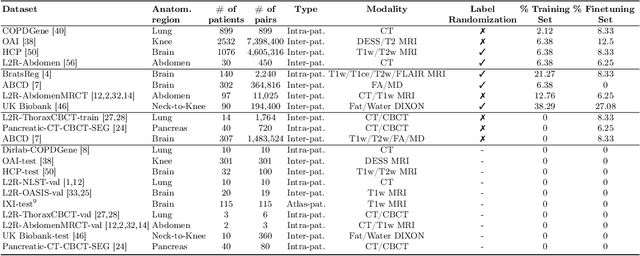
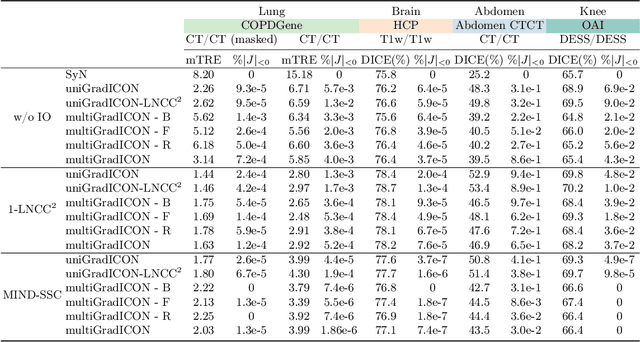
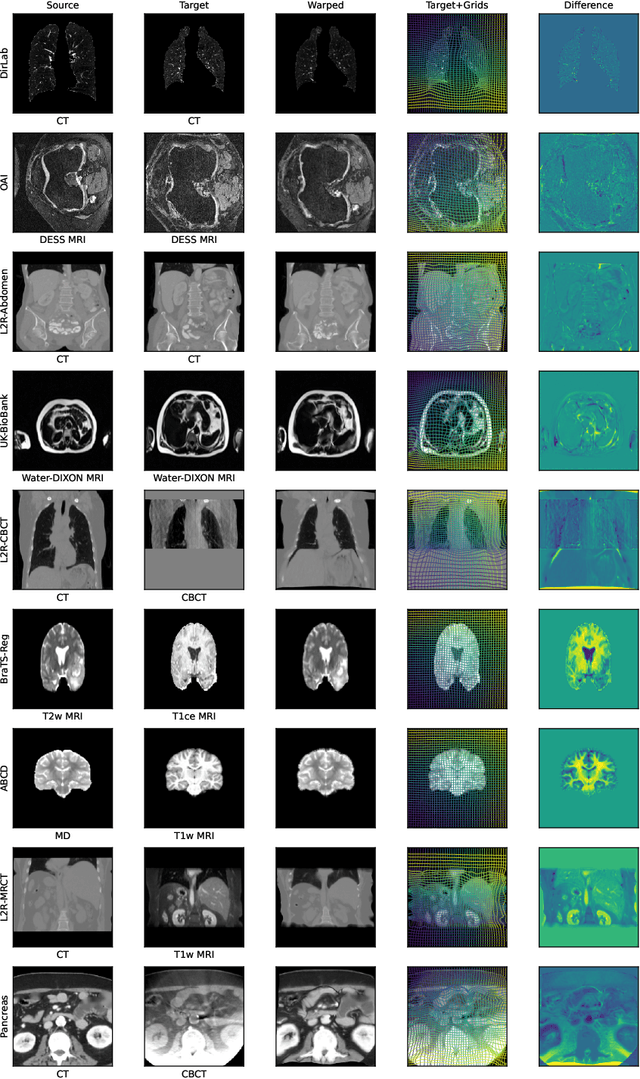
Abstract:Modern medical image registration approaches predict deformations using deep networks. These approaches achieve state-of-the-art (SOTA) registration accuracy and are generally fast. However, deep learning (DL) approaches are, in contrast to conventional non-deep-learning-based approaches, anatomy-specific. Recently, a universal deep registration approach, uniGradICON, has been proposed. However, uniGradICON focuses on monomodal image registration. In this work, we therefore develop multiGradICON as a first step towards universal *multimodal* medical image registration. Specifically, we show that 1) we can train a DL registration model that is suitable for monomodal *and* multimodal registration; 2) loss function randomization can increase multimodal registration accuracy; and 3) training a model with multimodal data helps multimodal generalization. Our code and the multiGradICON model are available at https://github.com/uncbiag/uniGradICON.
CARES: A Comprehensive Benchmark of Trustworthiness in Medical Vision Language Models
Jun 10, 2024
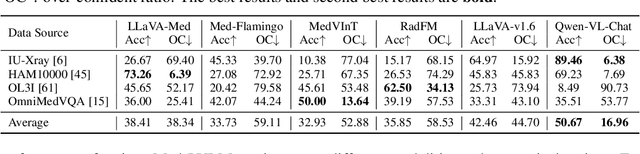
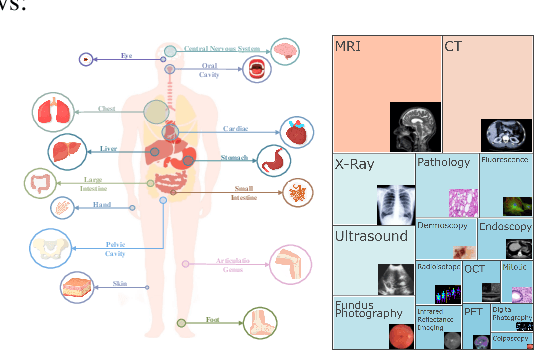
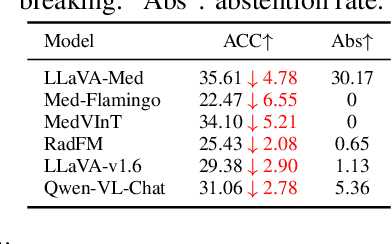
Abstract:Artificial intelligence has significantly impacted medical applications, particularly with the advent of Medical Large Vision Language Models (Med-LVLMs), sparking optimism for the future of automated and personalized healthcare. However, the trustworthiness of Med-LVLMs remains unverified, posing significant risks for future model deployment. In this paper, we introduce CARES and aim to comprehensively evaluate the Trustworthiness of Med-LVLMs across the medical domain. We assess the trustworthiness of Med-LVLMs across five dimensions, including trustfulness, fairness, safety, privacy, and robustness. CARES comprises about 41K question-answer pairs in both closed and open-ended formats, covering 16 medical image modalities and 27 anatomical regions. Our analysis reveals that the models consistently exhibit concerns regarding trustworthiness, often displaying factual inaccuracies and failing to maintain fairness across different demographic groups. Furthermore, they are vulnerable to attacks and demonstrate a lack of privacy awareness. We publicly release our benchmark and code in https://github.com/richard-peng-xia/CARES.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge