Raul San Jose Estepar
multiGradICON: A Foundation Model for Multimodal Medical Image Registration
Aug 01, 2024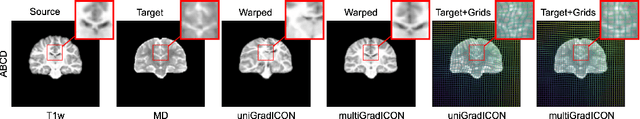
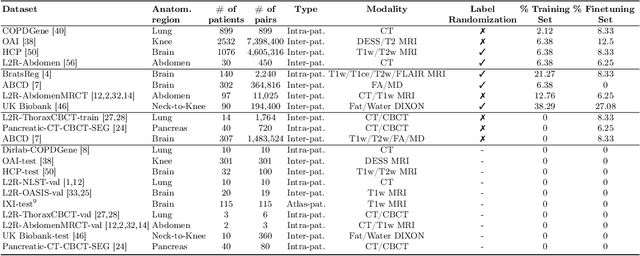
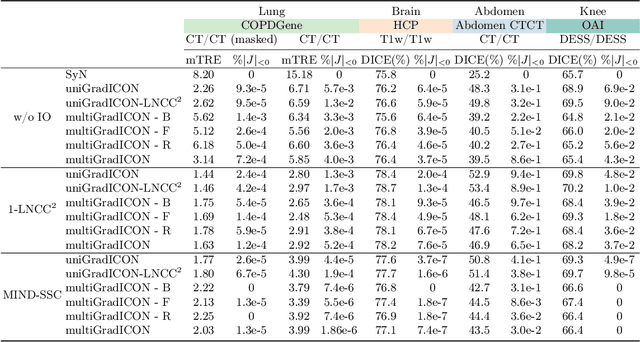
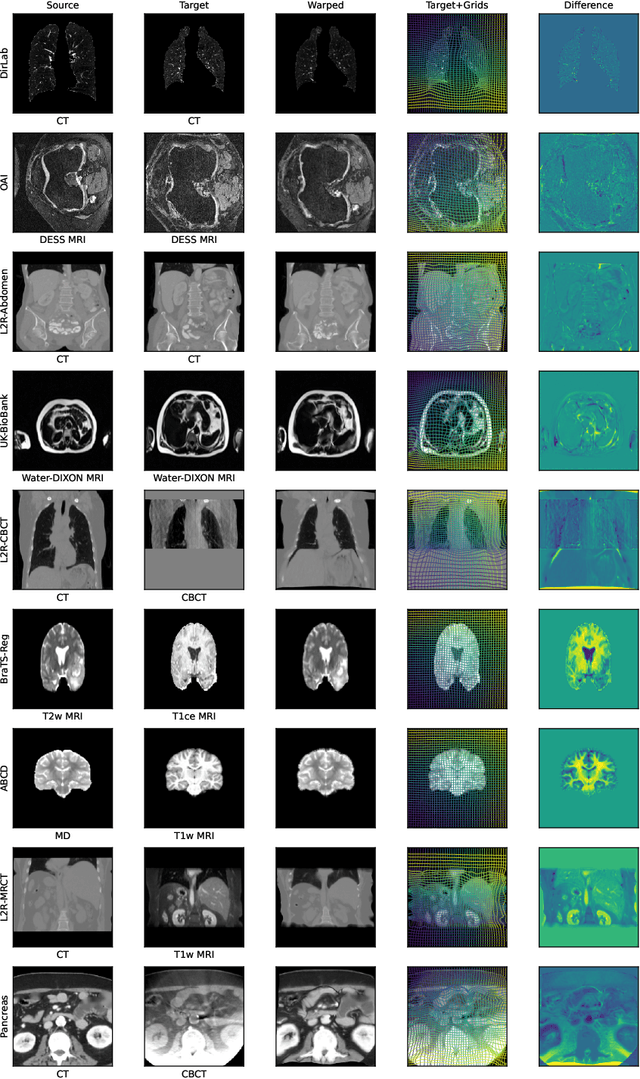
Abstract:Modern medical image registration approaches predict deformations using deep networks. These approaches achieve state-of-the-art (SOTA) registration accuracy and are generally fast. However, deep learning (DL) approaches are, in contrast to conventional non-deep-learning-based approaches, anatomy-specific. Recently, a universal deep registration approach, uniGradICON, has been proposed. However, uniGradICON focuses on monomodal image registration. In this work, we therefore develop multiGradICON as a first step towards universal *multimodal* medical image registration. Specifically, we show that 1) we can train a DL registration model that is suitable for monomodal *and* multimodal registration; 2) loss function randomization can increase multimodal registration accuracy; and 3) training a model with multimodal data helps multimodal generalization. Our code and the multiGradICON model are available at https://github.com/uncbiag/uniGradICON.
CARL: A Framework for Equivariant Image Registration
May 28, 2024
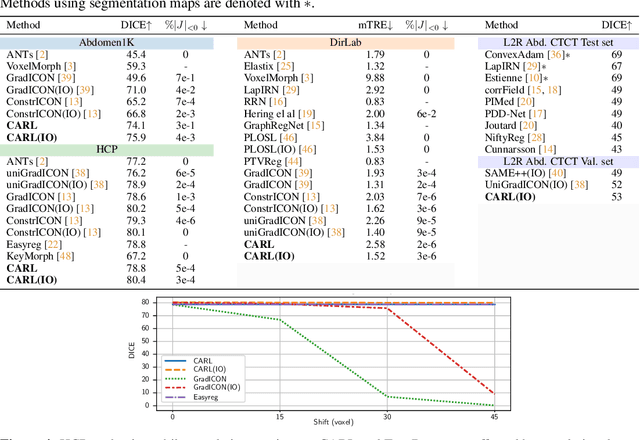
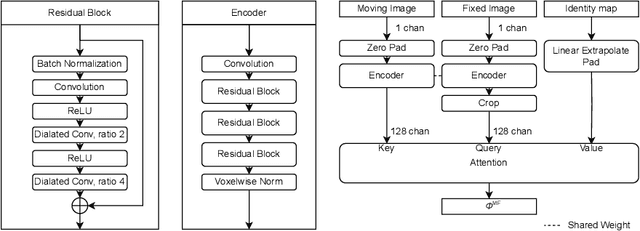
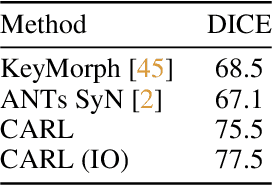
Abstract:Image registration estimates spatial correspondences between a pair of images. These estimates are typically obtained via numerical optimization or regression by a deep network. A desirable property of such estimators is that a correspondence estimate (e.g., the true oracle correspondence) for an image pair is maintained under deformations of the input images. Formally, the estimator should be equivariant to a desired class of image transformations. In this work, we present careful analyses of the desired equivariance properties in the context of multi-step deep registration networks. Based on these analyses we 1) introduce the notions of $[U,U]$ equivariance (network equivariance to the same deformations of the input images) and $[W,U]$ equivariance (where input images can undergo different deformations); we 2) show that in a suitable multi-step registration setup it is sufficient for overall $[W,U]$ equivariance if the first step has $[W,U]$ equivariance and all others have $[U,U]$ equivariance; we 3) show that common displacement-predicting networks only exhibit $[U,U]$ equivariance to translations instead of the more powerful $[W,U]$ equivariance; and we 4) show how to achieve multi-step $[W,U]$ equivariance via a coordinate-attention mechanism combined with displacement-predicting refinement layers (CARL). Overall, our approach obtains excellent practical registration performance on several 3D medical image registration tasks and outperforms existing unsupervised approaches for the challenging problem of abdomen registration.
uniGradICON: A Foundation Model for Medical Image Registration
Mar 09, 2024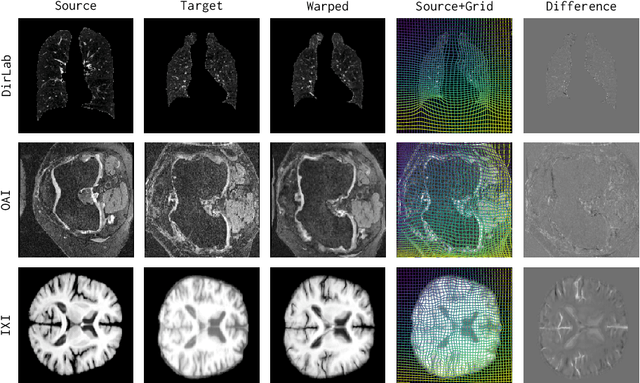
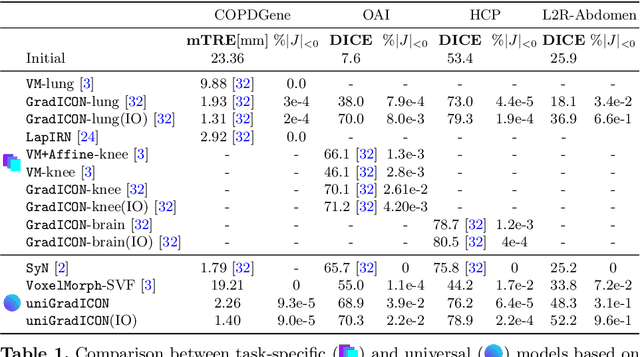

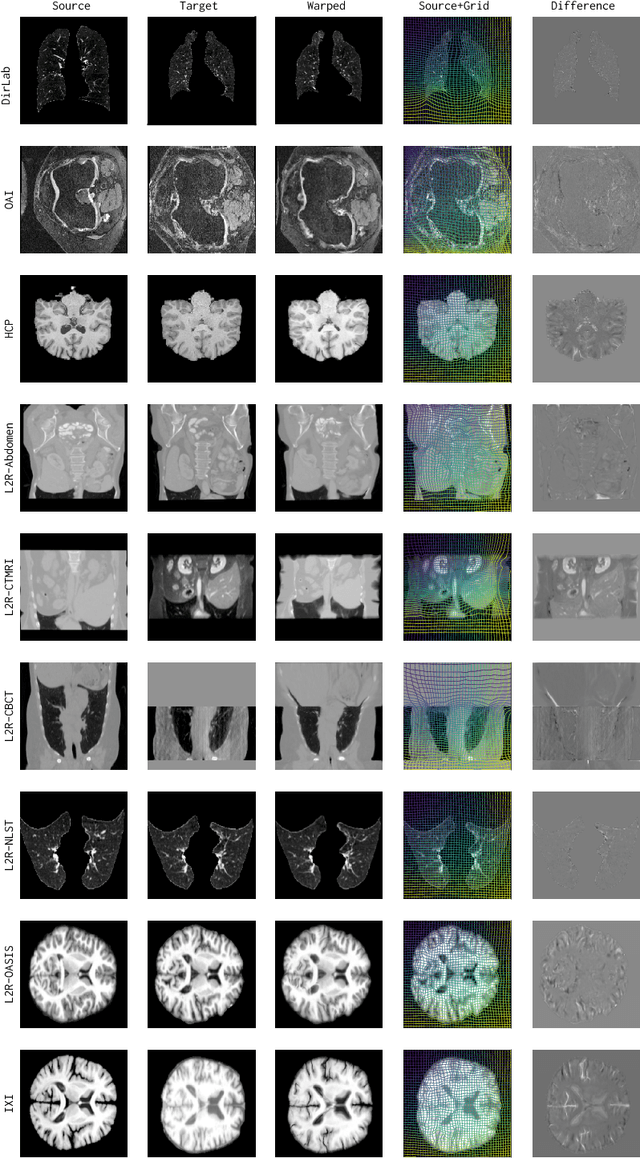
Abstract:Conventional medical image registration approaches directly optimize over the parameters of a transformation model. These approaches have been highly successful and are used generically for registrations of different anatomical regions. Recent deep registration networks are incredibly fast and accurate but are only trained for specific tasks. Hence, they are no longer generic registration approaches. We therefore propose uniGradICON, a first step toward a foundation model for registration providing 1) great performance \emph{across} multiple datasets which is not feasible for current learning-based registration methods, 2) zero-shot capabilities for new registration tasks suitable for different acquisitions, anatomical regions, and modalities compared to the training dataset, and 3) a strong initialization for finetuning on out-of-distribution registration tasks. UniGradICON unifies the speed and accuracy benefits of learning-based registration algorithms with the generic applicability of conventional non-deep-learning approaches. We extensively trained and evaluated uniGradICON on twelve different public datasets. Our code and the uniGradICON model are available at https://github.com/uncbiag/uniGradICON.
Inverse Consistency by Construction for Multistep Deep Registration
Apr 28, 2023Abstract:Inverse consistency is a desirable property for image registration. We propose a simple technique to make a neural registration network inverse consistent by construction, as a consequence of its structure, as long as it parameterizes its output transform by a Lie group. We extend this technique to multi-step neural registration by composing many such networks in a way that preserves inverse consistency. This multi-step approach also allows for inverse-consistent coarse to fine registration. We evaluate our technique on synthetic 2-D data and four 3-D medical image registration tasks and obtain excellent registration accuracy while assuring inverse consistency.
Multi-site, Multi-domain Airway Tree Modeling : A Public Benchmark for Pulmonary Airway Segmentation
Mar 10, 2023



Abstract:Open international challenges are becoming the de facto standard for assessing computer vision and image analysis algorithms. In recent years, new methods have extended the reach of pulmonary airway segmentation that is closer to the limit of image resolution. Since EXACT'09 pulmonary airway segmentation, limited effort has been directed to quantitative comparison of newly emerged algorithms driven by the maturity of deep learning based approaches and clinical drive for resolving finer details of distal airways for early intervention of pulmonary diseases. Thus far, public annotated datasets are extremely limited, hindering the development of data-driven methods and detailed performance evaluation of new algorithms. To provide a benchmark for the medical imaging community, we organized the Multi-site, Multi-domain Airway Tree Modeling (ATM'22), which was held as an official challenge event during the MICCAI 2022 conference. ATM'22 provides large-scale CT scans with detailed pulmonary airway annotation, including 500 CT scans (300 for training, 50 for validation, and 150 for testing). The dataset was collected from different sites and it further included a portion of noisy COVID-19 CTs with ground-glass opacity and consolidation. Twenty-three teams participated in the entire phase of the challenge and the algorithms for the top ten teams are reviewed in this paper. Quantitative and qualitative results revealed that deep learning models embedded with the topological continuity enhancement achieved superior performance in general. ATM'22 challenge holds as an open-call design, the training data and the gold standard evaluation are available upon successful registration via its homepage.
Accurate Point Cloud Registration with Robust Optimal Transport
Nov 01, 2021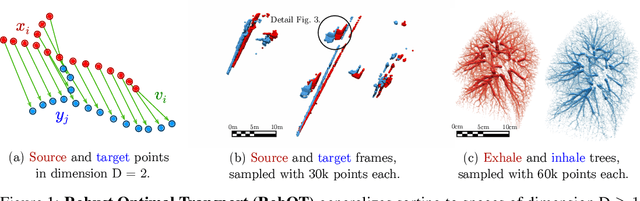
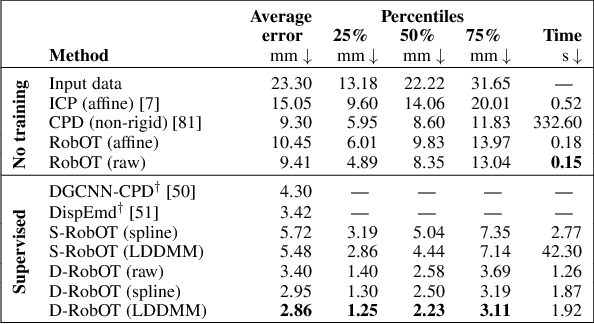
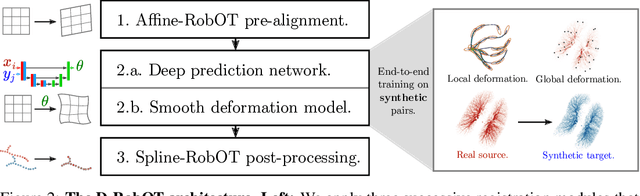
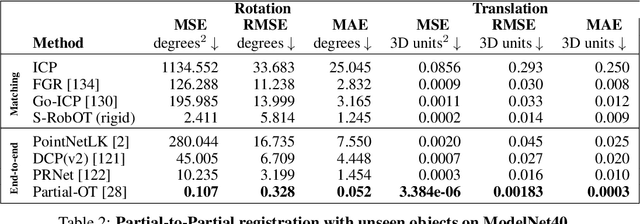
Abstract:This work investigates the use of robust optimal transport (OT) for shape matching. Specifically, we show that recent OT solvers improve both optimization-based and deep learning methods for point cloud registration, boosting accuracy at an affordable computational cost. This manuscript starts with a practical overview of modern OT theory. We then provide solutions to the main difficulties in using this framework for shape matching. Finally, we showcase the performance of transport-enhanced registration models on a wide range of challenging tasks: rigid registration for partial shapes; scene flow estimation on the Kitti dataset; and nonparametric registration of lung vascular trees between inspiration and expiration. Our OT-based methods achieve state-of-the-art results on Kitti and for the challenging lung registration task, both in terms of accuracy and scalability. We also release PVT1010, a new public dataset of 1,010 pairs of lung vascular trees with densely sampled points. This dataset provides a challenging use case for point cloud registration algorithms with highly complex shapes and deformations. Our work demonstrates that robust OT enables fast pre-alignment and fine-tuning for a wide range of registration models, thereby providing a new key method for the computer vision toolbox. Our code and dataset are available online at: https://github.com/uncbiag/robot.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge