Chenhao Pei
Multi-site, Multi-domain Airway Tree Modeling : A Public Benchmark for Pulmonary Airway Segmentation
Mar 10, 2023



Abstract:Open international challenges are becoming the de facto standard for assessing computer vision and image analysis algorithms. In recent years, new methods have extended the reach of pulmonary airway segmentation that is closer to the limit of image resolution. Since EXACT'09 pulmonary airway segmentation, limited effort has been directed to quantitative comparison of newly emerged algorithms driven by the maturity of deep learning based approaches and clinical drive for resolving finer details of distal airways for early intervention of pulmonary diseases. Thus far, public annotated datasets are extremely limited, hindering the development of data-driven methods and detailed performance evaluation of new algorithms. To provide a benchmark for the medical imaging community, we organized the Multi-site, Multi-domain Airway Tree Modeling (ATM'22), which was held as an official challenge event during the MICCAI 2022 conference. ATM'22 provides large-scale CT scans with detailed pulmonary airway annotation, including 500 CT scans (300 for training, 50 for validation, and 150 for testing). The dataset was collected from different sites and it further included a portion of noisy COVID-19 CTs with ground-glass opacity and consolidation. Twenty-three teams participated in the entire phase of the challenge and the algorithms for the top ten teams are reviewed in this paper. Quantitative and qualitative results revealed that deep learning models embedded with the topological continuity enhancement achieved superior performance in general. ATM'22 challenge holds as an open-call design, the training data and the gold standard evaluation are available upon successful registration via its homepage.
Brain Tumor Segmentation Network Using Attention-based Fusion and Spatial Relationship Constraint
Oct 31, 2020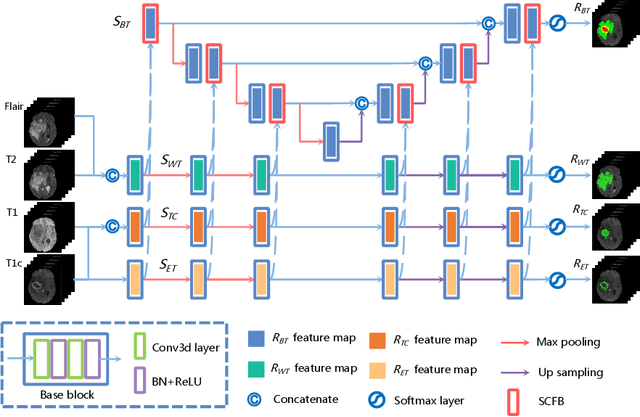
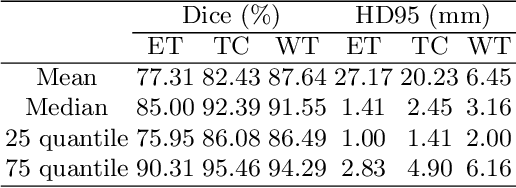
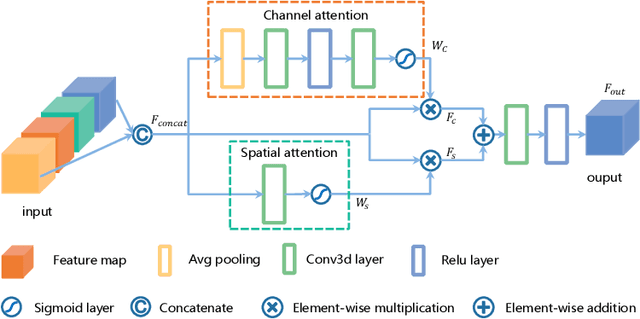
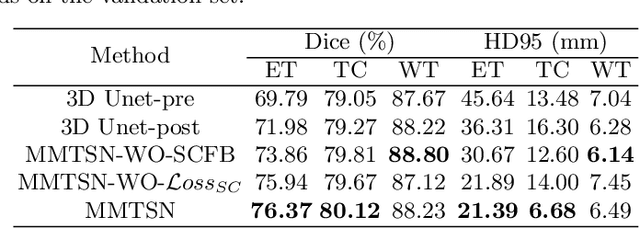
Abstract:Delineating the brain tumor from magnetic resonance (MR) images is critical for the treatment of gliomas. However, automatic delineation is challenging due to the complex appearance and ambiguous outlines of tumors. Considering that multi-modal MR images can reflect different tumor biological properties, we develop a novel multi-modal tumor segmentation network (MMTSN) to robustly segment brain tumors based on multi-modal MR images. The MMTSN is composed of three sub-branches and a main branch. Specifically, the sub-branches are used to capture different tumor features from multi-modal images, while in the main branch, we design a spatial-channel fusion block (SCFB) to effectively aggregate multi-modal features. Additionally, inspired by the fact that the spatial relationship between sub-regions of tumor is relatively fixed, e.g., the enhancing tumor is always in the tumor core, we propose a spatial loss to constrain the relationship between different sub-regions of tumor. We evaluate our method on the test set of multi-modal brain tumor segmentation challenge 2020 (BraTs2020). The method achieves 0.8764, 0.8243 and 0.773 dice score for whole tumor, tumor core and enhancing tumor, respectively.
Multi-Modality Pathology Segmentation Framework: Application to Cardiac Magnetic Resonance Images
Aug 13, 2020



Abstract:Multi-sequence of cardiac magnetic resonance (CMR) images can provide complementary information for myocardial pathology (scar and edema). However, it is still challenging to fuse these underlying information for pathology segmentation effectively. This work presents an automatic cascade pathology segmentation framework based on multi-modality CMR images. It mainly consists of two neural networks: an anatomical structure segmentation network (ASSN) and a pathological region segmentation network (PRSN). Specifically, the ASSN aims to segment the anatomical structure where the pathology may exist, and it can provide a spatial prior for the pathological region segmentation. In addition, we integrate a denoising auto-encoder (DAE) into the ASSN to generate segmentation results with plausible shapes. The PRSN is designed to segment pathological region based on the result of ASSN, in which a fusion block based on channel attention is proposed to better aggregate multi-modality information from multi-modality CMR images. Experiments from the MyoPS2020 challenge dataset show that our framework can achieve promising performance for myocardial scar and edema segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge