Pengxin Yu
Hunting imaging biomarkers in pulmonary fibrosis: Benchmarks of the AIIB23 challenge
Dec 21, 2023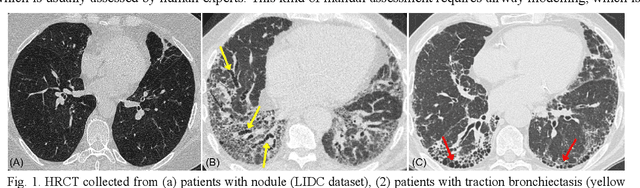
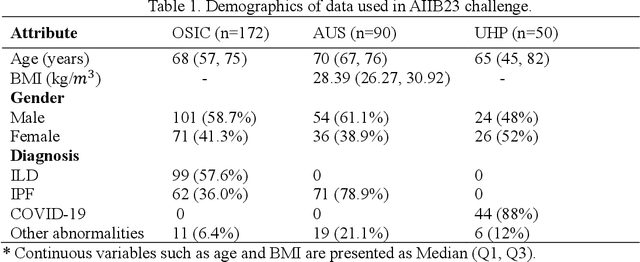
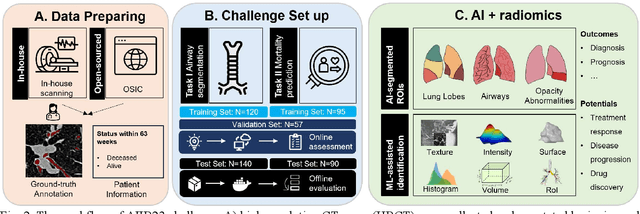
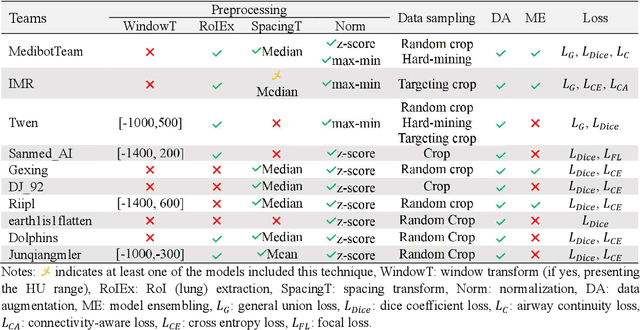
Abstract:Airway-related quantitative imaging biomarkers are crucial for examination, diagnosis, and prognosis in pulmonary diseases. However, the manual delineation of airway trees remains prohibitively time-consuming. While significant efforts have been made towards enhancing airway modelling, current public-available datasets concentrate on lung diseases with moderate morphological variations. The intricate honeycombing patterns present in the lung tissues of fibrotic lung disease patients exacerbate the challenges, often leading to various prediction errors. To address this issue, the 'Airway-Informed Quantitative CT Imaging Biomarker for Fibrotic Lung Disease 2023' (AIIB23) competition was organized in conjunction with the official 2023 International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI). The airway structures were meticulously annotated by three experienced radiologists. Competitors were encouraged to develop automatic airway segmentation models with high robustness and generalization abilities, followed by exploring the most correlated QIB of mortality prediction. A training set of 120 high-resolution computerised tomography (HRCT) scans were publicly released with expert annotations and mortality status. The online validation set incorporated 52 HRCT scans from patients with fibrotic lung disease and the offline test set included 140 cases from fibrosis and COVID-19 patients. The results have shown that the capacity of extracting airway trees from patients with fibrotic lung disease could be enhanced by introducing voxel-wise weighted general union loss and continuity loss. In addition to the competitive image biomarkers for prognosis, a strong airway-derived biomarker (Hazard ratio>1.5, p<0.0001) was revealed for survival prognostication compared with existing clinical measurements, clinician assessment and AI-based biomarkers.
Efficient automatic segmentation for multi-level pulmonary arteries: The PARSE challenge
Apr 07, 2023



Abstract:Efficient automatic segmentation of multi-level (i.e. main and branch) pulmonary arteries (PA) in CTPA images plays a significant role in clinical applications. However, most existing methods concentrate only on main PA or branch PA segmentation separately and ignore segmentation efficiency. Besides, there is no public large-scale dataset focused on PA segmentation, which makes it highly challenging to compare the different methods. To benchmark multi-level PA segmentation algorithms, we organized the first \textbf{P}ulmonary \textbf{AR}tery \textbf{SE}gmentation (PARSE) challenge. On the one hand, we focus on both the main PA and the branch PA segmentation. On the other hand, for better clinical application, we assign the same score weight to segmentation efficiency (mainly running time and GPU memory consumption during inference) while ensuring PA segmentation accuracy. We present a summary of the top algorithms and offer some suggestions for efficient and accurate multi-level PA automatic segmentation. We provide the PARSE challenge as open-access for the community to benchmark future algorithm developments at \url{https://parse2022.grand-challenge.org/Parse2022/}.
Multi-site, Multi-domain Airway Tree Modeling : A Public Benchmark for Pulmonary Airway Segmentation
Mar 10, 2023



Abstract:Open international challenges are becoming the de facto standard for assessing computer vision and image analysis algorithms. In recent years, new methods have extended the reach of pulmonary airway segmentation that is closer to the limit of image resolution. Since EXACT'09 pulmonary airway segmentation, limited effort has been directed to quantitative comparison of newly emerged algorithms driven by the maturity of deep learning based approaches and clinical drive for resolving finer details of distal airways for early intervention of pulmonary diseases. Thus far, public annotated datasets are extremely limited, hindering the development of data-driven methods and detailed performance evaluation of new algorithms. To provide a benchmark for the medical imaging community, we organized the Multi-site, Multi-domain Airway Tree Modeling (ATM'22), which was held as an official challenge event during the MICCAI 2022 conference. ATM'22 provides large-scale CT scans with detailed pulmonary airway annotation, including 500 CT scans (300 for training, 50 for validation, and 150 for testing). The dataset was collected from different sites and it further included a portion of noisy COVID-19 CTs with ground-glass opacity and consolidation. Twenty-three teams participated in the entire phase of the challenge and the algorithms for the top ten teams are reviewed in this paper. Quantitative and qualitative results revealed that deep learning models embedded with the topological continuity enhancement achieved superior performance in general. ATM'22 challenge holds as an open-call design, the training data and the gold standard evaluation are available upon successful registration via its homepage.
MMMNA-Net for Overall Survival Time Prediction of Brain Tumor Patients
Jun 13, 2022



Abstract:Overall survival (OS) time is one of the most important evaluation indices for gliomas situations. Multimodal Magnetic Resonance Imaging (MRI) scans play an important role in the study of glioma prognosis OS time. Several deep learning-based methods are proposed for the OS time prediction on multi-modal MRI problems. However, these methods usually fuse multi-modal information at the beginning or at the end of the deep learning networks and lack the fusion of features from different scales. In addition, the fusion at the end of networks always adapts global with global (eg. fully connected after concatenation of global average pooling output) or local with local (eg. bilinear pooling), which loses the information of local with global. In this paper, we propose a novel method for multi-modal OS time prediction of brain tumor patients, which contains an improved nonlocal features fusion module introduced on different scales. Our method obtains a relative 8.76% improvement over the current state-of-art method (0.6989 vs. 0.6426 on accuracy). Extensive testing demonstrates that our method could adapt to situations with missing modalities. The code is available at https://github.com/TangWen920812/mmmna-net.
RPLHR-CT Dataset and Transformer Baseline for Volumetric Super-Resolution from CT Scans
Jun 13, 2022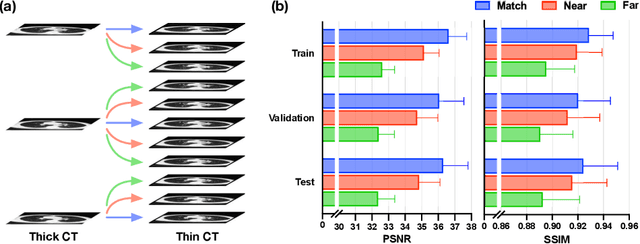
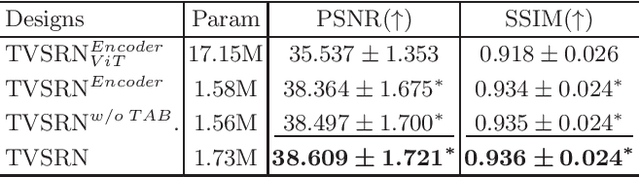
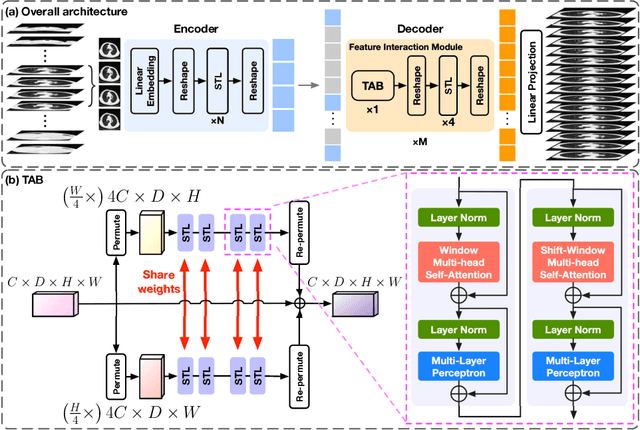
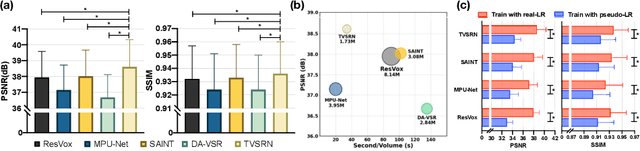
Abstract:In clinical practice, anisotropic volumetric medical images with low through-plane resolution are commonly used due to short acquisition time and lower storage cost. Nevertheless, the coarse resolution may lead to difficulties in medical diagnosis by either physicians or computer-aided diagnosis algorithms. Deep learning-based volumetric super-resolution (SR) methods are feasible ways to improve resolution, with convolutional neural networks (CNN) at their core. Despite recent progress, these methods are limited by inherent properties of convolution operators, which ignore content relevance and cannot effectively model long-range dependencies. In addition, most of the existing methods use pseudo-paired volumes for training and evaluation, where pseudo low-resolution (LR) volumes are generated by a simple degradation of their high-resolution (HR) counterparts. However, the domain gap between pseudo- and real-LR volumes leads to the poor performance of these methods in practice. In this paper, we build the first public real-paired dataset RPLHR-CT as a benchmark for volumetric SR, and provide baseline results by re-implementing four state-of-the-art CNN-based methods. Considering the inherent shortcoming of CNN, we also propose a transformer volumetric super-resolution network (TVSRN) based on attention mechanisms, dispensing with convolutions entirely. This is the first research to use a pure transformer for CT volumetric SR. The experimental results show that TVSRN significantly outperforms all baselines on both PSNR and SSIM. Moreover, the TVSRN method achieves a better trade-off between the image quality, the number of parameters, and the running time. Data and code are available at https://github.com/smilenaxx/RPLHR-CT.
Transformer Lesion Tracker
Jun 13, 2022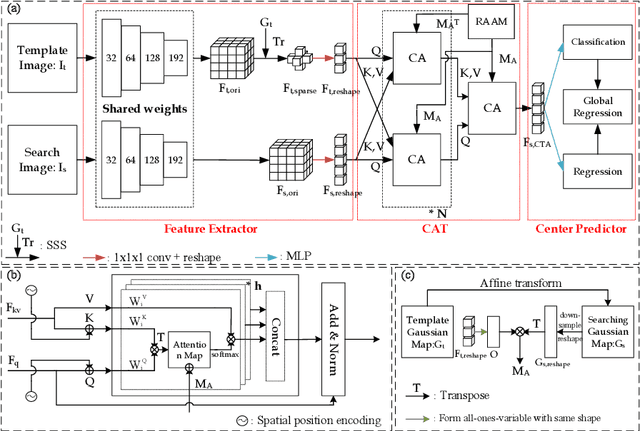
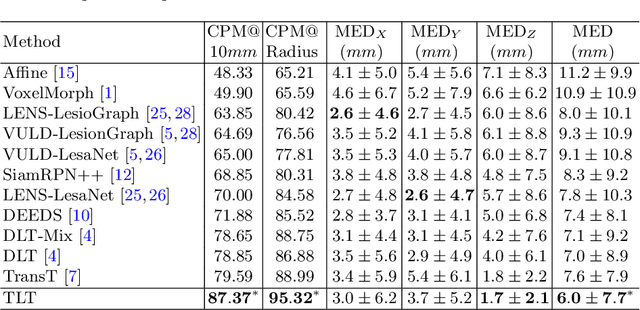
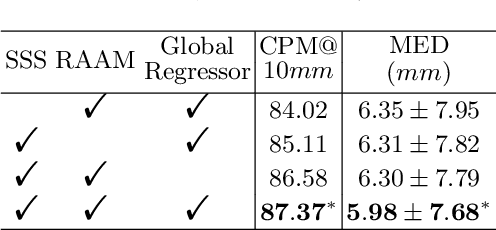
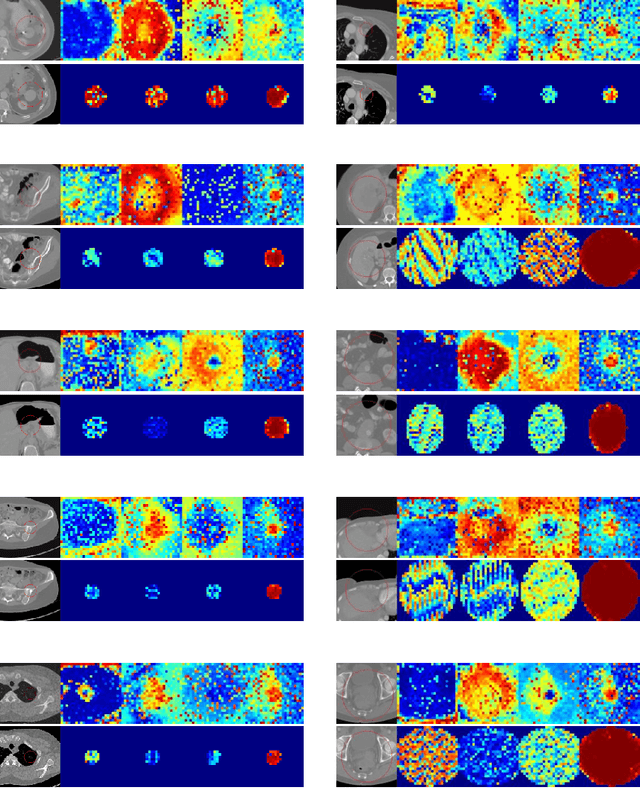
Abstract:Evaluating lesion progression and treatment response via longitudinal lesion tracking plays a critical role in clinical practice. Automated approaches for this task are motivated by prohibitive labor costs and time consumption when lesion matching is done manually. Previous methods typically lack the integration of local and global information. In this work, we propose a transformer-based approach, termed Transformer Lesion Tracker (TLT). Specifically, we design a Cross Attention-based Transformer (CAT) to capture and combine both global and local information to enhance feature extraction. We also develop a Registration-based Anatomical Attention Module (RAAM) to introduce anatomical information to CAT so that it can focus on useful feature knowledge. A Sparse Selection Strategy (SSS) is presented for selecting features and reducing memory footprint in Transformer training. In addition, we use a global regression to further improve model performance. We conduct experiments on a public dataset to show the superiority of our method and find that our model performance has improved the average Euclidean center error by at least 14.3% (6mm vs. 7mm) compared with the state-of-the-art (SOTA). Code is available at https://github.com/TangWen920812/TLT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge