Corey W. Arnold
Computational Mapping of Reactive Stroma in Prostate Cancer Yields Interpretable, Prognostic Biomarkers
Jan 10, 2026Abstract:Current histopathological grading of prostate cancer relies primarily on glandular architecture, largely overlooking the tumor microenvironment. Here, we present PROTAS, a deep learning framework that quantifies reactive stroma (RS) in routine hematoxylin and eosin (H&E) slides and links stromal morphology to underlying biology. PROTAS-defined RS is characterized by nuclear enlargement, collagen disorganization, and transcriptomic enrichment of contractile pathways. PROTAS detects RS robustly in the external Prostate, Lung, Colorectal, and Ovarian (PLCO) dataset and, using domain-adversarial training, generalizes to diagnostic biopsies. In head-to-head comparisons, PROTAS outperforms pathologists for RS detection, and spatial RS features predict biochemical recurrence independently of established prognostic variables (c-index 0.80). By capturing subtle stromal phenotypes associated with tumor progression, PROTAS provides an interpretable, scalable biomarker to refine risk stratification.
Zero-shot Medical Event Prediction Using a Generative Pre-trained Transformer on Electronic Health Records
Mar 07, 2025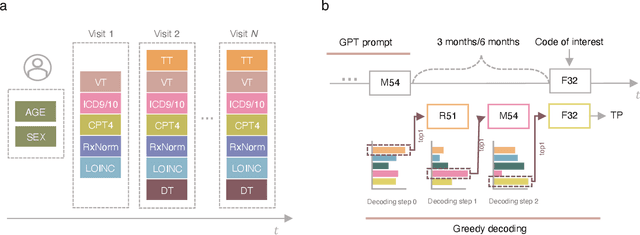
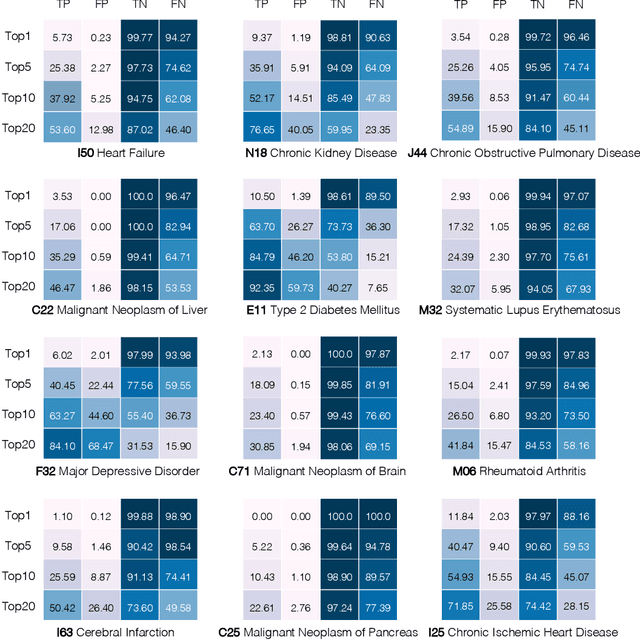
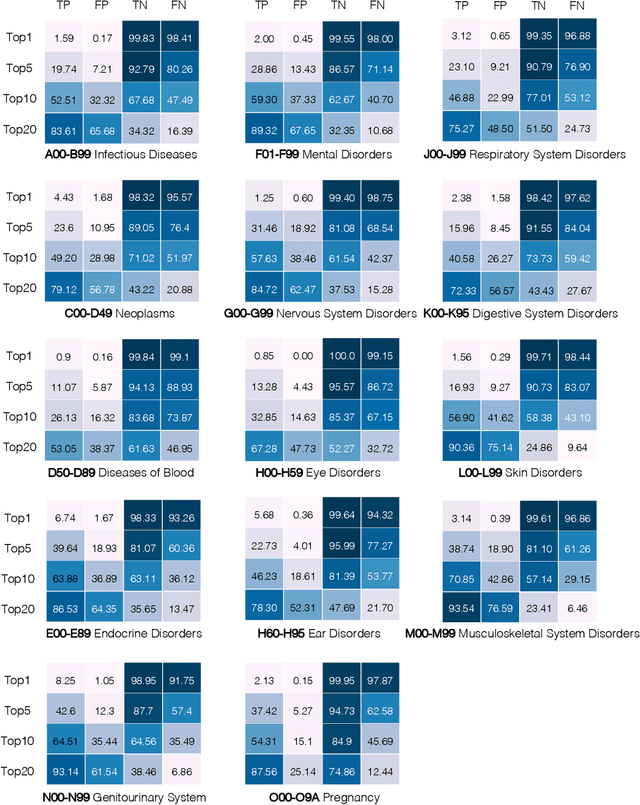
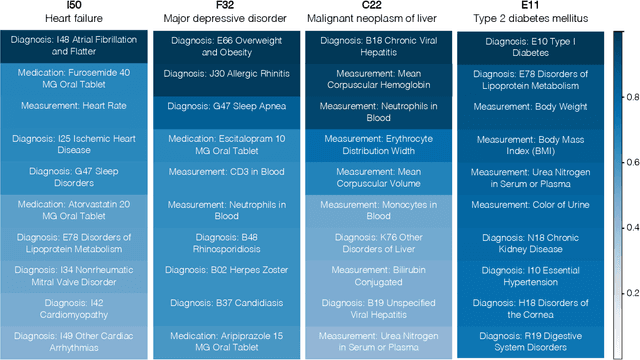
Abstract:Longitudinal data in electronic health records (EHRs) represent an individual`s clinical history through a sequence of codified concepts, including diagnoses, procedures, medications, and laboratory tests. Foundational models, such as generative pre-trained transformers (GPT), can leverage this data to predict future events. While fine-tuning of these models enhances task-specific performance, it is costly, complex, and unsustainable for every target. We show that a foundation model trained on EHRs can perform predictive tasks in a zero-shot manner, eliminating the need for fine-tuning. This study presents the first comprehensive analysis of zero-shot forecasting with GPT-based foundational models in EHRs, introducing a novel pipeline that formulates medical concept prediction as a generative modeling task. Unlike supervised approaches requiring extensive labeled data, our method enables the model to forecast a next medical event purely from a pretraining knowledge. We evaluate performance across multiple time horizons and clinical categories, demonstrating model`s ability to capture latent temporal dependencies and complex patient trajectories without task supervision. Model performance for predicting the next medical concept was evaluated using precision and recall metrics, achieving an average top1 precision of 0.614 and recall of 0.524. For 12 major diagnostic conditions, the model demonstrated strong zero-shot performance, achieving high true positive rates while maintaining low false positives. We demonstrate the power of a foundational EHR GPT model in capturing diverse phenotypes and enabling robust, zero-shot forecasting of clinical outcomes. This capability enhances the versatility of predictive healthcare models and reduces the need for task-specific training, enabling more scalable applications in clinical settings.
Reducing Overtreatment of Indeterminate Thyroid Nodules Using a Multimodal Deep Learning Model
Sep 27, 2024
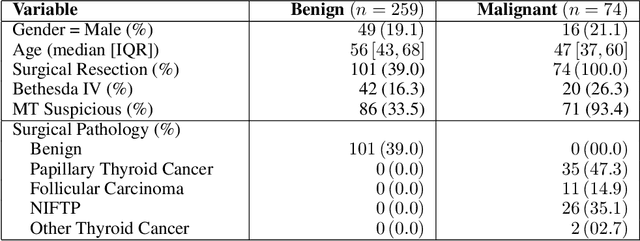
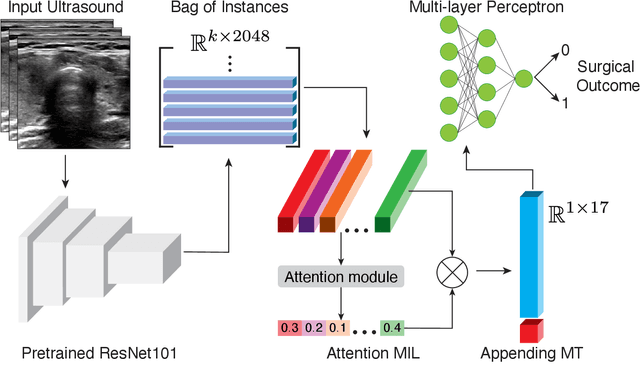
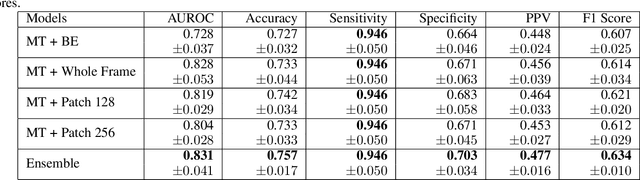
Abstract:Objective: Molecular testing (MT) classifies cytologically indeterminate thyroid nodules as benign or malignant with high sensitivity but low positive predictive value (PPV), only using molecular profiles, ignoring ultrasound (US) imaging and biopsy. We address this limitation by applying attention multiple instance learning (AMIL) to US images. Methods: We retrospectively reviewed 333 patients with indeterminate thyroid nodules at UCLA medical center (259 benign, 74 malignant). A multi-modal deep learning AMIL model was developed, combining US images and MT to classify the nodules as benign or malignant and enhance the malignancy risk stratification of MT. Results: The final AMIL model matched MT sensitivity (0.946) while significantly improving PPV (0.477 vs 0.448 for MT alone), indicating fewer false positives while maintaining high sensitivity. Conclusion: Our approach reduces false positives compared to MT while maintaining the same ability to identify positive cases, potentially reducing unnecessary benign thyroid resections in patients with indeterminate nodules.
Digital Volumetric Biopsy Cores Improve Gleason Grading of Prostate Cancer Using Deep Learning
Sep 12, 2024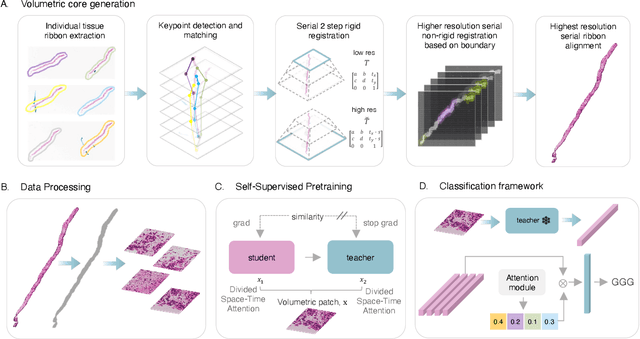
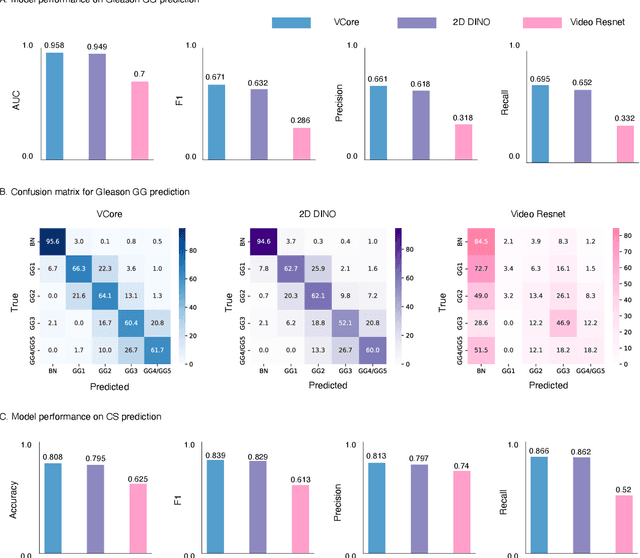
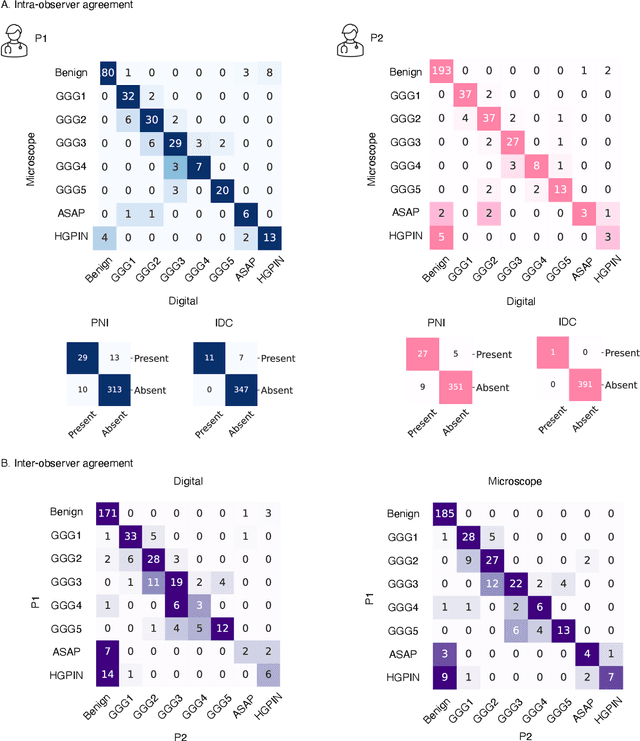

Abstract:Prostate cancer (PCa) was the most frequently diagnosed cancer among American men in 2023. The histological grading of biopsies is essential for diagnosis, and various deep learning-based solutions have been developed to assist with this task. Existing deep learning frameworks are typically applied to individual 2D cross-sections sliced from 3D biopsy tissue specimens. This process impedes the analysis of complex tissue structures such as glands, which can vary depending on the tissue slice examined. We propose a novel digital pathology data source called a "volumetric core," obtained via the extraction and co-alignment of serially sectioned tissue sections using a novel morphology-preserving alignment framework. We trained an attention-based multiple-instance learning (ABMIL) framework on deep features extracted from volumetric patches to automatically classify the Gleason Grade Group (GGG). To handle volumetric patches, we used a modified video transformer with a deep feature extractor pretrained using self-supervised learning. We ran our morphology-preserving alignment framework to construct 10,210 volumetric cores, leaving out 30% for pretraining. The rest of the dataset was used to train ABMIL, which resulted in a 0.958 macro-average AUC, 0.671 F1 score, 0.661 precision, and 0.695 recall averaged across all five GGG significantly outperforming the 2D baselines.
Ultrasound Image Enhancement using CycleGAN and Perceptual Loss
Dec 18, 2023Abstract:Purpose: The objective of this work is to introduce an advanced framework designed to enhance ultrasound images, especially those captured by portable hand-held devices, which often produce lower quality images due to hardware constraints. Additionally, this framework is uniquely capable of effectively handling non-registered input ultrasound image pairs, addressing a common challenge in medical imaging. Materials and Methods: In this retrospective study, we utilized an enhanced generative adversarial network (CycleGAN) model for ultrasound image enhancement across five organ systems. Perceptual loss, derived from deep features of pretrained neural networks, is applied to ensure the human-perceptual quality of the enhanced images. These images are compared with paired images acquired from high resolution devices to demonstrate the model's ability to generate realistic high-quality images across organ systems. Results: Preliminary validation of the framework reveals promising performance metrics. The model generates images that result in a Structural Similarity Index (SSI) score of 0.722, Locally Normalized Cross-Correlation (LNCC) score of 0.902 and 28.802 for the Peak Signal-to-Noise Ratio (PSNR) metric. Conclusion: This work presents a significant advancement in medical imaging through the development of a CycleGAN model enhanced with Perceptual Loss (PL), effectively bridging the quality gap between ultrasound images from varied devices. By training on paired images, the model not only improves image quality but also ensures the preservation of vital anatomic structural content. This approach may improve equity in access to healthcare by enhancing portable device capabilities, although further validation and optimizations are necessary for broader clinical application.
Predicting Thrombectomy Recanalization from CT Imaging Using Deep Learning Models
Feb 08, 2023Abstract:For acute ischemic stroke (AIS) patients with large vessel occlusions, clinicians must decide if the benefit of mechanical thrombectomy (MTB) outweighs the risks and potential complications following an invasive procedure. Pre-treatment computed tomography (CT) and angiography (CTA) are widely used to characterize occlusions in the brain vasculature. If a patient is deemed eligible, a modified treatment in cerebral ischemia (mTICI) score will be used to grade how well blood flow is reestablished throughout and following the MTB procedure. An estimation of the likelihood of successful recanalization can support treatment decision-making. In this study, we proposed a fully automated prediction of a patient's recanalization score using pre-treatment CT and CTA imaging. We designed a spatial cross attention network (SCANet) that utilizes vision transformers to localize to pertinent slices and brain regions. Our top model achieved an average cross-validated ROC-AUC of 77.33 $\pm$ 3.9\%. This is a promising result that supports future applications of deep learning on CT and CTA for the identification of eligible AIS patients for MTB.
* Medical Imaging with Deep Learning 2022 accepted short paper Jun 2022
RPLHR-CT Dataset and Transformer Baseline for Volumetric Super-Resolution from CT Scans
Jun 13, 2022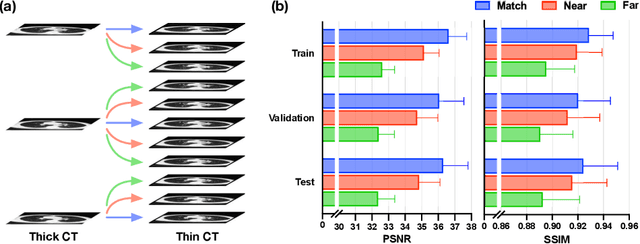
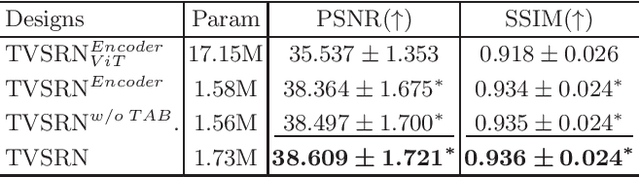
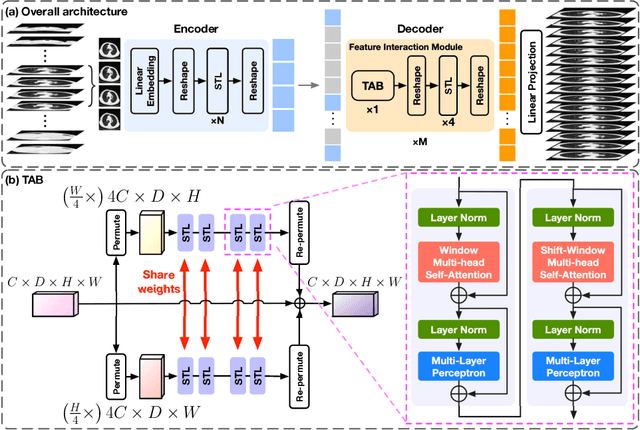
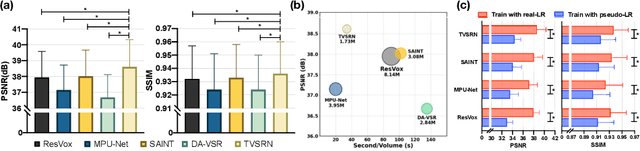
Abstract:In clinical practice, anisotropic volumetric medical images with low through-plane resolution are commonly used due to short acquisition time and lower storage cost. Nevertheless, the coarse resolution may lead to difficulties in medical diagnosis by either physicians or computer-aided diagnosis algorithms. Deep learning-based volumetric super-resolution (SR) methods are feasible ways to improve resolution, with convolutional neural networks (CNN) at their core. Despite recent progress, these methods are limited by inherent properties of convolution operators, which ignore content relevance and cannot effectively model long-range dependencies. In addition, most of the existing methods use pseudo-paired volumes for training and evaluation, where pseudo low-resolution (LR) volumes are generated by a simple degradation of their high-resolution (HR) counterparts. However, the domain gap between pseudo- and real-LR volumes leads to the poor performance of these methods in practice. In this paper, we build the first public real-paired dataset RPLHR-CT as a benchmark for volumetric SR, and provide baseline results by re-implementing four state-of-the-art CNN-based methods. Considering the inherent shortcoming of CNN, we also propose a transformer volumetric super-resolution network (TVSRN) based on attention mechanisms, dispensing with convolutions entirely. This is the first research to use a pure transformer for CT volumetric SR. The experimental results show that TVSRN significantly outperforms all baselines on both PSNR and SSIM. Moreover, the TVSRN method achieves a better trade-off between the image quality, the number of parameters, and the running time. Data and code are available at https://github.com/smilenaxx/RPLHR-CT.
Transformer Lesion Tracker
Jun 13, 2022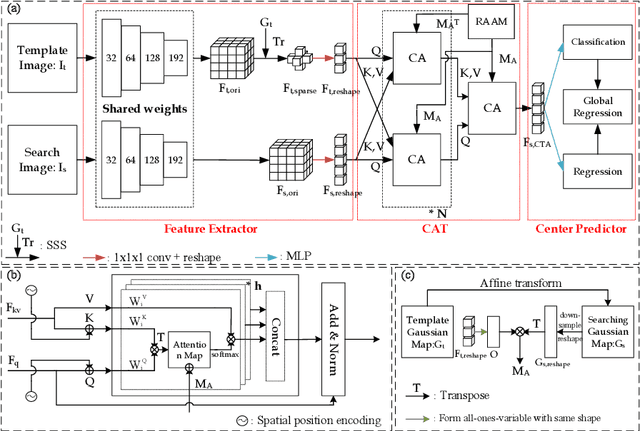
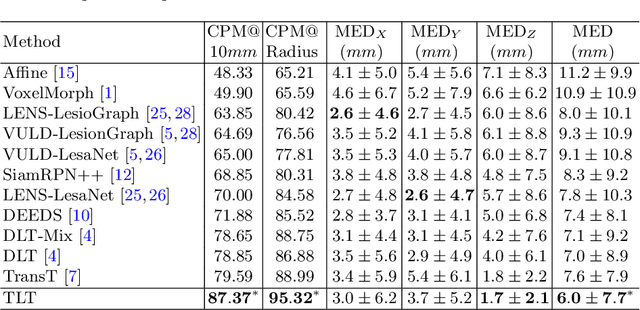
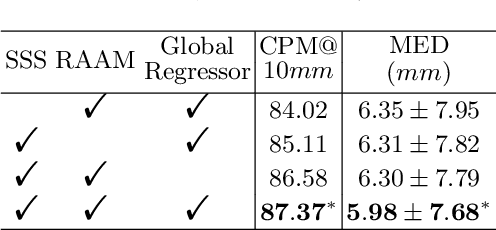
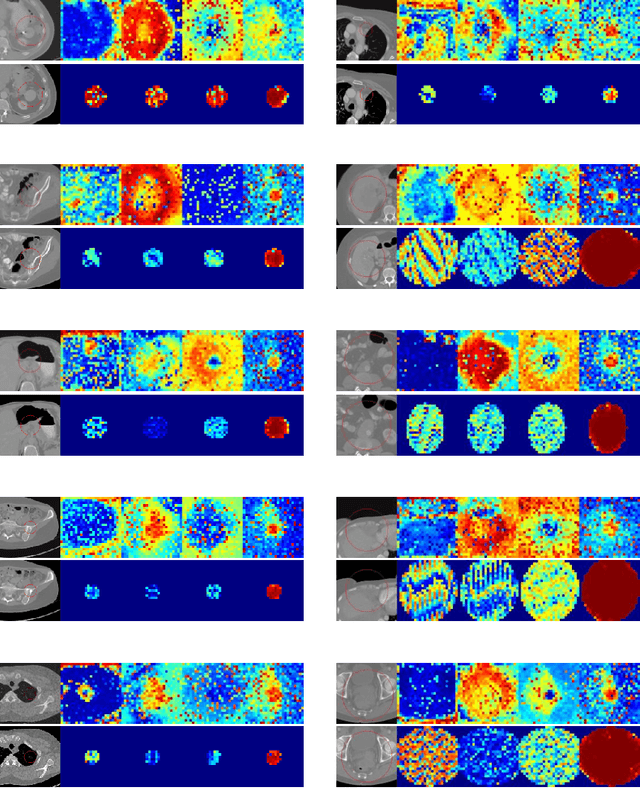
Abstract:Evaluating lesion progression and treatment response via longitudinal lesion tracking plays a critical role in clinical practice. Automated approaches for this task are motivated by prohibitive labor costs and time consumption when lesion matching is done manually. Previous methods typically lack the integration of local and global information. In this work, we propose a transformer-based approach, termed Transformer Lesion Tracker (TLT). Specifically, we design a Cross Attention-based Transformer (CAT) to capture and combine both global and local information to enhance feature extraction. We also develop a Registration-based Anatomical Attention Module (RAAM) to introduce anatomical information to CAT so that it can focus on useful feature knowledge. A Sparse Selection Strategy (SSS) is presented for selecting features and reducing memory footprint in Transformer training. In addition, we use a global regression to further improve model performance. We conduct experiments on a public dataset to show the superiority of our method and find that our model performance has improved the average Euclidean center error by at least 14.3% (6mm vs. 7mm) compared with the state-of-the-art (SOTA). Code is available at https://github.com/TangWen920812/TLT.
Federated Learning with Research Prototypes for Multi-Center MRI-based Detection of Prostate Cancer with Diverse Histopathology
Jun 11, 2022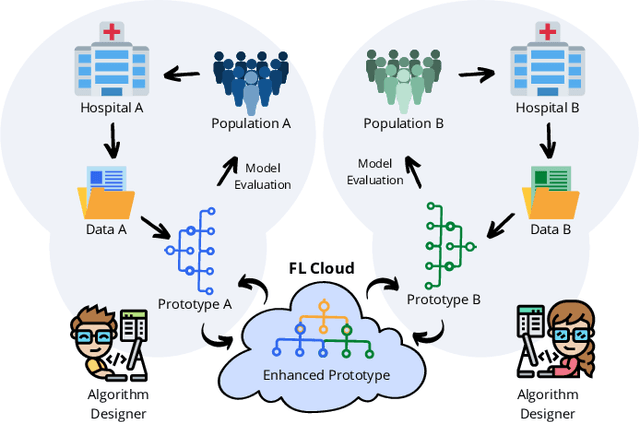
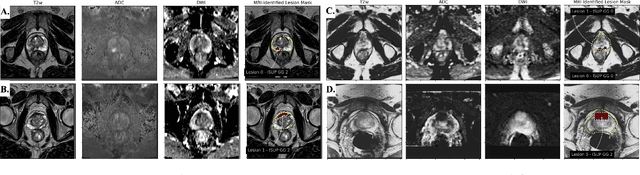
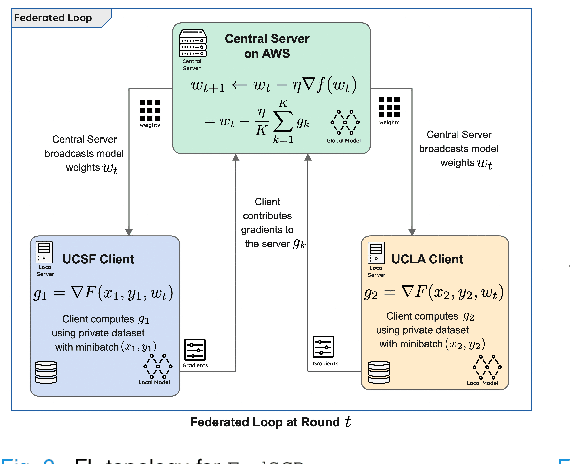
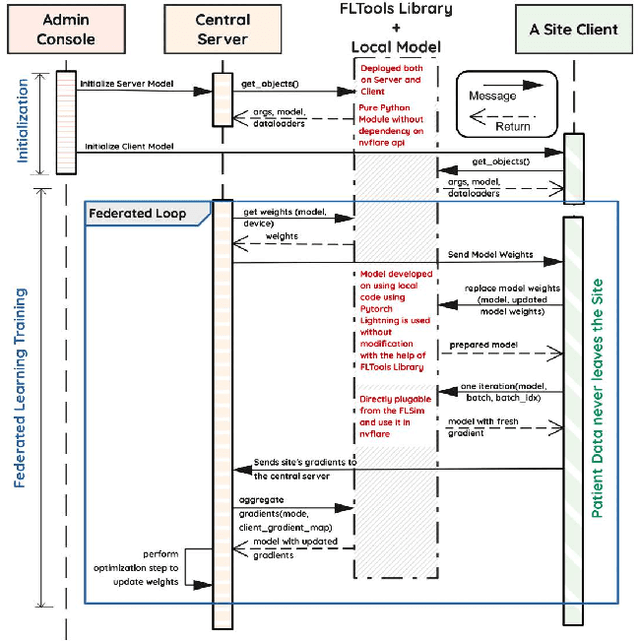
Abstract:Early prostate cancer detection and staging from MRI are extremely challenging tasks for both radiologists and deep learning algorithms, but the potential to learn from large and diverse datasets remains a promising avenue to increase their generalization capability both within- and across clinics. To enable this for prototype-stage algorithms, where the majority of existing research remains, in this paper we introduce a flexible federated learning framework for cross-site training, validation, and evaluation of deep prostate cancer detection algorithms. Our approach utilizes an abstracted representation of the model architecture and data, which allows unpolished prototype deep learning models to be trained without modification using the NVFlare federated learning framework. Our results show increases in prostate cancer detection and classification accuracy using a specialized neural network model and diverse prostate biopsy data collected at two University of California research hospitals, demonstrating the efficacy of our approach in adapting to different datasets and improving MR-biomarker discovery. We open-source our FLtools system, which can be easily adapted to other deep learning projects for medical imaging.
Graph Convolutional Networks for Multi-modality Medical Imaging: Methods, Architectures, and Clinical Applications
Feb 17, 2022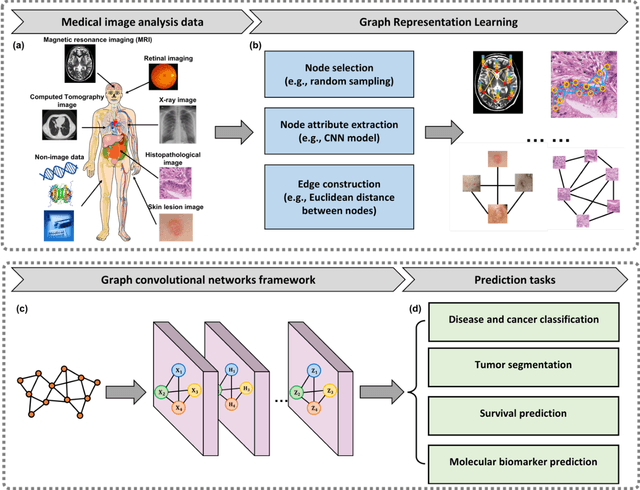
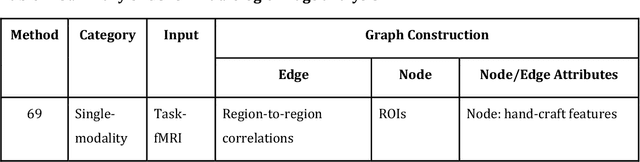
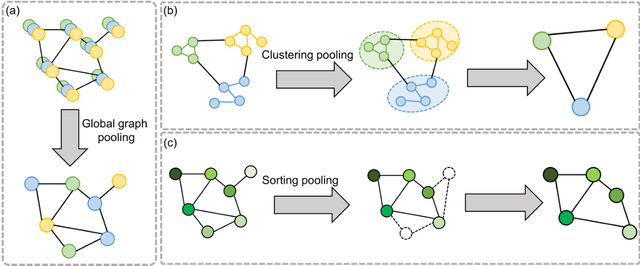
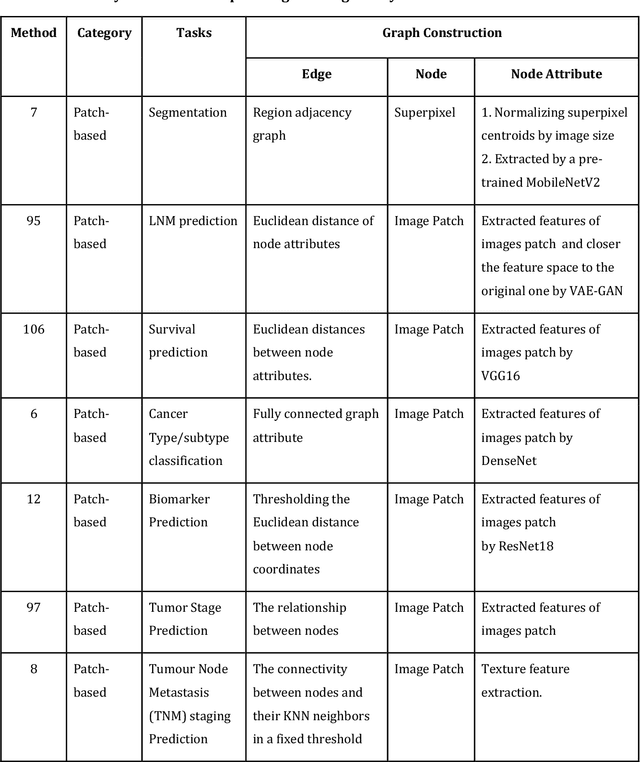
Abstract:Image-based characterization and disease understanding involve integrative analysis of morphological, spatial, and topological information across biological scales. The development of graph convolutional networks (GCNs) has created the opportunity to address this information complexity via graph-driven architectures, since GCNs can perform feature aggregation, interaction, and reasoning with remarkable flexibility and efficiency. These GCNs capabilities have spawned a new wave of research in medical imaging analysis with the overarching goal of improving quantitative disease understanding, monitoring, and diagnosis. Yet daunting challenges remain for designing the important image-to-graph transformation for multi-modality medical imaging and gaining insights into model interpretation and enhanced clinical decision support. In this review, we present recent GCNs developments in the context of medical image analysis including imaging data from radiology and histopathology. We discuss the fast-growing use of graph network architectures in medical image analysis to improve disease diagnosis and patient outcomes in clinical practice. To foster cross-disciplinary research, we present GCNs technical advancements, emerging medical applications, identify common challenges in the use of image-based GCNs and their extensions in model interpretation, large-scale benchmarks that promise to transform the scope of medical image studies and related graph-driven medical research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge