Kexin Ding
MedForge: Building Medical Foundation Models Like Open Source Software Development
Feb 22, 2025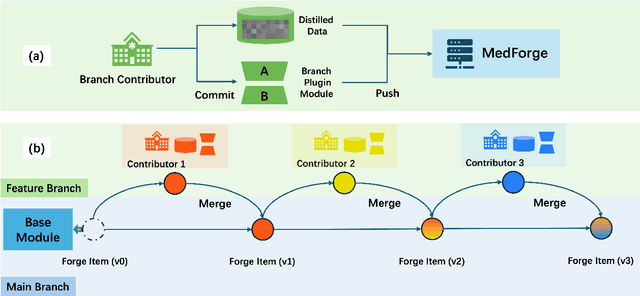
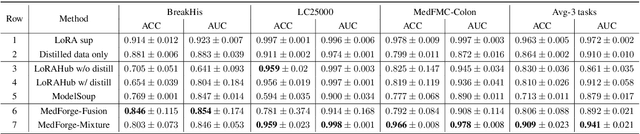
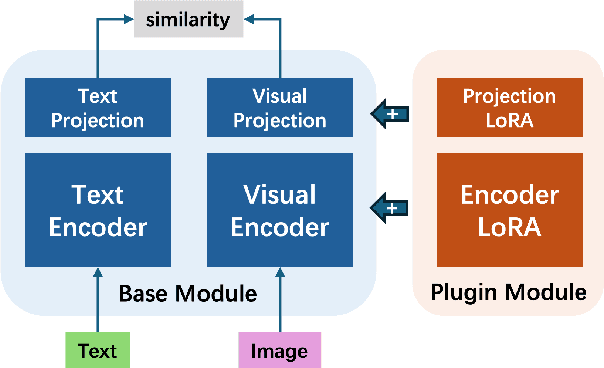
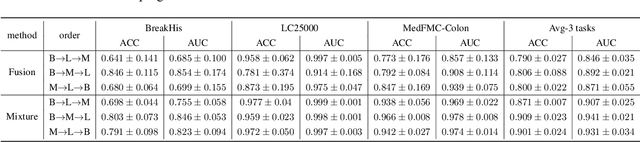
Abstract:Foundational models (FMs) have made significant strides in the healthcare domain. Yet the data silo challenge and privacy concern remain in healthcare systems, hindering safe medical data sharing and collaborative model development among institutions. The collection and curation of scalable clinical datasets increasingly become the bottleneck for training strong FMs. In this study, we propose Medical Foundation Models Merging (MedForge), a cooperative framework enabling a community-driven medical foundation model development, meanwhile preventing the information leakage of raw patient data and mitigating synchronization model development issues across clinical institutions. MedForge offers a bottom-up model construction mechanism by flexibly merging task-specific Low-Rank Adaptation (LoRA) modules, which can adapt to downstream tasks while retaining original model parameters. Through an asynchronous LoRA module integration scheme, the resulting composite model can progressively enhance its comprehensive performance on various clinical tasks. MedForge shows strong performance on multiple clinical datasets (e.g., breast cancer, lung cancer, and colon cancer) collected from different institutions. Our major findings highlight the value of collaborative foundation models in advancing multi-center clinical collaboration effectively and cohesively. Our code is publicly available at https://github.com/TanZheling/MedForge.
Data-Centric Foundation Models in Computational Healthcare: A Survey
Jan 04, 2024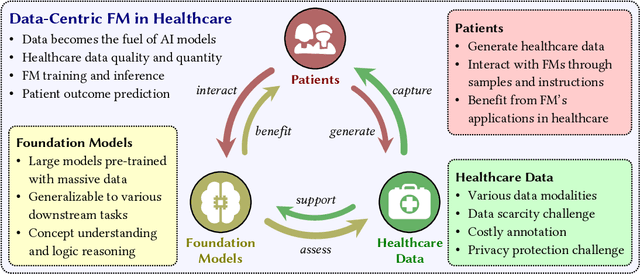
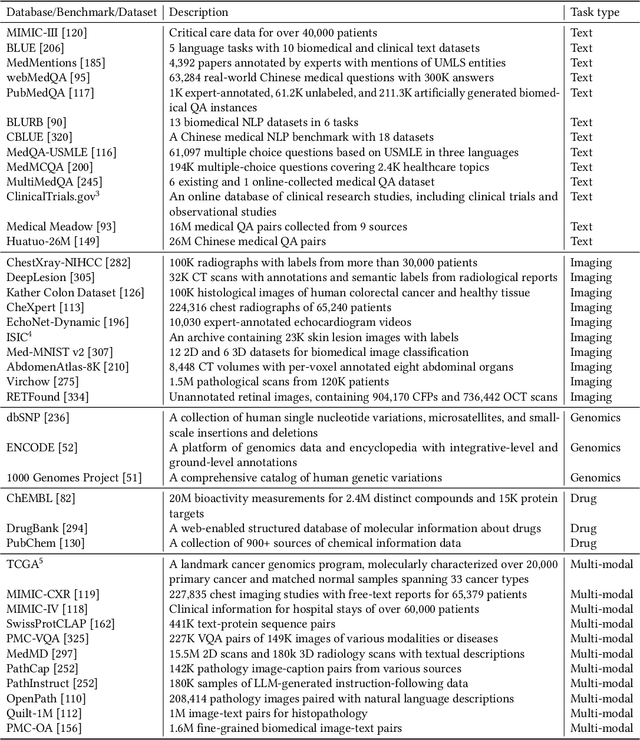
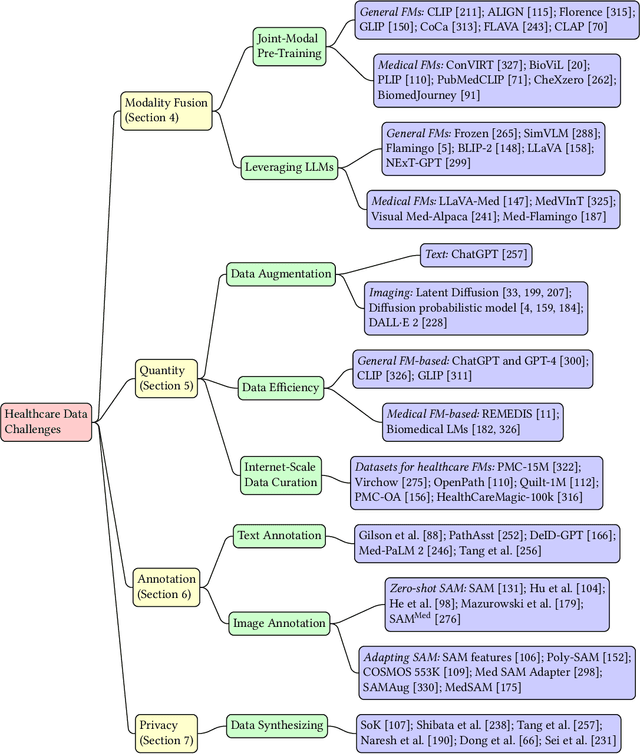
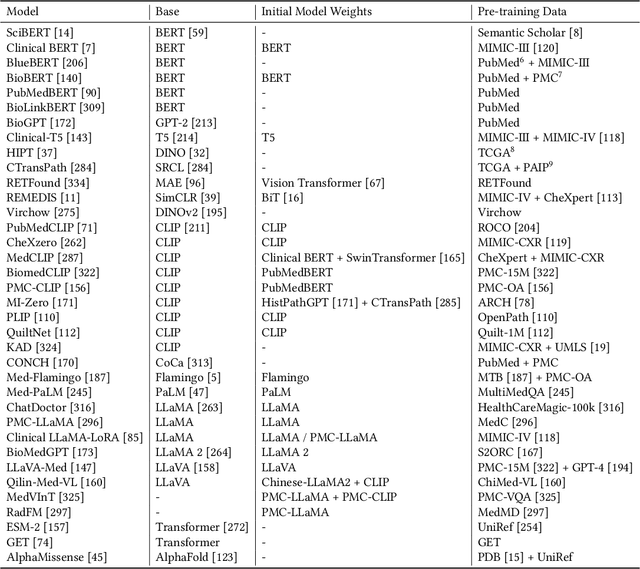
Abstract:The advent of foundation models (FMs) as an emerging suite of AI techniques has struck a wave of opportunities in computational healthcare. The interactive nature of these models, guided by pre-training data and human instructions, has ignited a data-centric AI paradigm that emphasizes better data characterization, quality, and scale. In healthcare AI, obtaining and processing high-quality clinical data records has been a longstanding challenge, ranging from data quantity, annotation, patient privacy, and ethics. In this survey, we investigate a wide range of data-centric approaches in the FM era (from model pre-training to inference) towards improving the healthcare workflow. We discuss key perspectives in AI security, assessment, and alignment with human values. Finally, we offer a promising outlook of FM-based analytics to enhance the performance of patient outcome and clinical workflow in the evolving landscape of healthcare and medicine. We provide an up-to-date list of healthcare-related foundation models and datasets at https://github.com/Yunkun-Zhang/Data-Centric-FM-Healthcare .
Pathology-and-genomics Multimodal Transformer for Survival Outcome Prediction
Jul 22, 2023


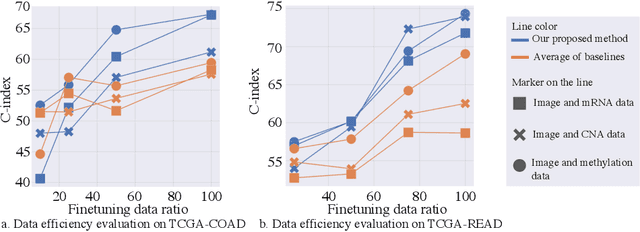
Abstract:Survival outcome assessment is challenging and inherently associated with multiple clinical factors (e.g., imaging and genomics biomarkers) in cancer. Enabling multimodal analytics promises to reveal novel predictive patterns of patient outcomes. In this study, we propose a multimodal transformer (PathOmics) integrating pathology and genomics insights into colon-related cancer survival prediction. We emphasize the unsupervised pretraining to capture the intrinsic interaction between tissue microenvironments in gigapixel whole slide images (WSIs) and a wide range of genomics data (e.g., mRNA-sequence, copy number variant, and methylation). After the multimodal knowledge aggregation in pretraining, our task-specific model finetuning could expand the scope of data utility applicable to both multi- and single-modal data (e.g., image- or genomics-only). We evaluate our approach on both TCGA colon and rectum cancer cohorts, showing that the proposed approach is competitive and outperforms state-of-the-art studies. Finally, our approach is desirable to utilize the limited number of finetuned samples towards data-efficient analytics for survival outcome prediction. The code is available at https://github.com/Cassie07/PathOmics.
Graph Convolutional Networks for Multi-modality Medical Imaging: Methods, Architectures, and Clinical Applications
Feb 17, 2022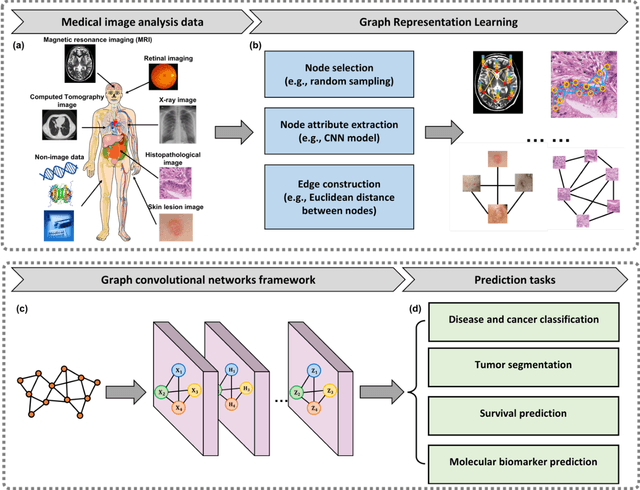
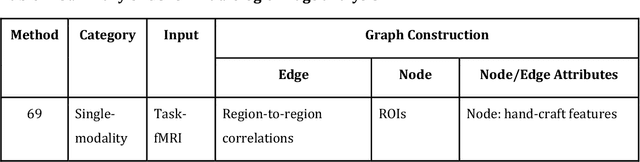
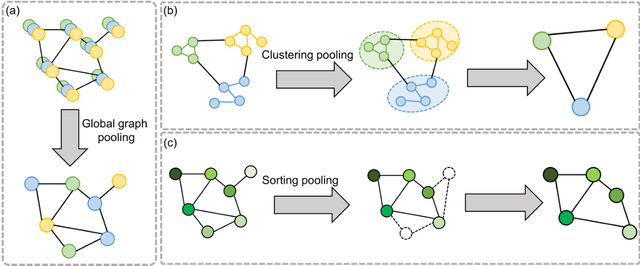
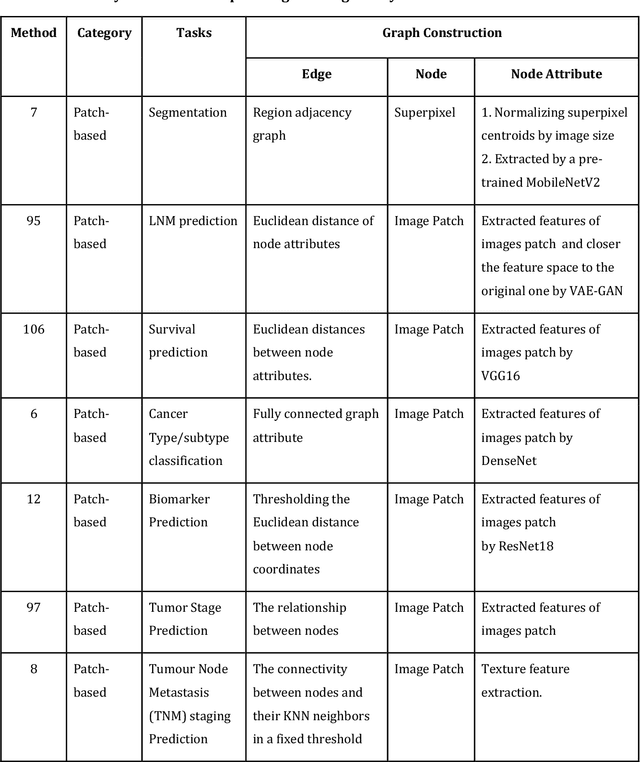
Abstract:Image-based characterization and disease understanding involve integrative analysis of morphological, spatial, and topological information across biological scales. The development of graph convolutional networks (GCNs) has created the opportunity to address this information complexity via graph-driven architectures, since GCNs can perform feature aggregation, interaction, and reasoning with remarkable flexibility and efficiency. These GCNs capabilities have spawned a new wave of research in medical imaging analysis with the overarching goal of improving quantitative disease understanding, monitoring, and diagnosis. Yet daunting challenges remain for designing the important image-to-graph transformation for multi-modality medical imaging and gaining insights into model interpretation and enhanced clinical decision support. In this review, we present recent GCNs developments in the context of medical image analysis including imaging data from radiology and histopathology. We discuss the fast-growing use of graph network architectures in medical image analysis to improve disease diagnosis and patient outcomes in clinical practice. To foster cross-disciplinary research, we present GCNs technical advancements, emerging medical applications, identify common challenges in the use of image-based GCNs and their extensions in model interpretation, large-scale benchmarks that promise to transform the scope of medical image studies and related graph-driven medical research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge