Dimitris Metaxas
Data Augmentation for High-Fidelity Generation of CAR-T/NK Immunological Synapse Images
Feb 03, 2026Abstract:Chimeric antigen receptor (CAR)-T and NK cell immunotherapies have transformed cancer treatment, and recent studies suggest that the quality of the CAR-T/NK cell immunological synapse (IS) may serve as a functional biomarker for predicting therapeutic efficacy. Accurate detection and segmentation of CAR-T/NK IS structures using artificial neural networks (ANNs) can greatly increase the speed and reliability of IS quantification. However, a persistent challenge is the limited size of annotated microscopy datasets, which restricts the ability of ANNs to generalize. To address this challenge, we integrate two complementary data-augmentation frameworks. First, we employ Instance Aware Automatic Augmentation (IAAA), an automated, instance-preserving augmentation method that generates synthetic CAR-T/NK IS images and corresponding segmentation masks by applying optimized augmentation policies to original IS data. IAAA supports multiple imaging modalities (e.g., fluorescence and brightfield) and can be applied directly to CAR-T/NK IS images derived from patient samples. In parallel, we introduce a Semantic-Aware AI Augmentation (SAAA) pipeline that combines a diffusion-based mask generator with a Pix2Pix conditional image synthesizer. This second method enables the creation of diverse, anatomically realistic segmentation masks and produces high-fidelity CAR-T/NK IS images aligned with those masks, further expanding the training corpus beyond what IAAA alone can provide. Together, these augmentation strategies generate synthetic images whose visual and structural properties closely match real IS data, significantly improving CAR-T/NK IS detection and segmentation performance. By enhancing the robustness and accuracy of IS quantification, this work supports the development of more reliable imaging-based biomarkers for predicting patient response to CAR-T/NK immunotherapy.
Anatomy-VLM: A Fine-grained Vision-Language Model for Medical Interpretation
Nov 11, 2025Abstract:Accurate disease interpretation from radiology remains challenging due to imaging heterogeneity. Achieving expert-level diagnostic decisions requires integration of subtle image features with clinical knowledge. Yet major vision-language models (VLMs) treat images as holistic entities and overlook fine-grained image details that are vital for disease diagnosis. Clinicians analyze images by utilizing their prior medical knowledge and identify anatomical structures as important region of interests (ROIs). Inspired from this human-centric workflow, we introduce Anatomy-VLM, a fine-grained, vision-language model that incorporates multi-scale information. First, we design a model encoder to localize key anatomical features from entire medical images. Second, these regions are enriched with structured knowledge for contextually-aware interpretation. Finally, the model encoder aligns multi-scale medical information to generate clinically-interpretable disease prediction. Anatomy-VLM achieves outstanding performance on both in- and out-of-distribution datasets. We also validate the performance of Anatomy-VLM on downstream image segmentation tasks, suggesting that its fine-grained alignment captures anatomical and pathology-related knowledge. Furthermore, the Anatomy-VLM's encoder facilitates zero-shot anatomy-wise interpretation, providing its strong expert-level clinical interpretation capabilities.
AutoEdit: Automatic Hyperparameter Tuning for Image Editing
Sep 18, 2025Abstract:Recent advances in diffusion models have revolutionized text-guided image editing, yet existing editing methods face critical challenges in hyperparameter identification. To get the reasonable editing performance, these methods often require the user to brute-force tune multiple interdependent hyperparameters, such as inversion timesteps and attention modification, \textit{etc.} This process incurs high computational costs due to the huge hyperparameter search space. We consider searching optimal editing's hyperparameters as a sequential decision-making task within the diffusion denoising process. Specifically, we propose a reinforcement learning framework, which establishes a Markov Decision Process that dynamically adjusts hyperparameters across denoising steps, integrating editing objectives into a reward function. The method achieves time efficiency through proximal policy optimization while maintaining optimal hyperparameter configurations. Experiments demonstrate significant reduction in search time and computational overhead compared to existing brute-force approaches, advancing the practical deployment of a diffusion-based image editing framework in the real world.
Token-Level Uncertainty Estimation for Large Language Model Reasoning
May 16, 2025Abstract:While Large Language Models (LLMs) have demonstrated impressive capabilities, their output quality remains inconsistent across various application scenarios, making it difficult to identify trustworthy responses, especially in complex tasks requiring multi-step reasoning. In this paper, we propose a token-level uncertainty estimation framework to enable LLMs to self-assess and self-improve their generation quality in mathematical reasoning. Specifically, we introduce low-rank random weight perturbation to LLM decoding, generating predictive distributions that we use to estimate token-level uncertainties. We then aggregate these uncertainties to reflect semantic uncertainty of the generated sequences. Experiments on mathematical reasoning datasets of varying difficulty demonstrate that our token-level uncertainty metrics strongly correlate with answer correctness and model robustness. Additionally, we explore using uncertainty to directly enhance the model's reasoning performance through multiple generations and the particle filtering algorithm. Our approach consistently outperforms existing uncertainty estimation methods, establishing effective uncertainty estimation as a valuable tool for both evaluating and improving reasoning generation in LLMs.
MedForge: Building Medical Foundation Models Like Open Source Software Development
Feb 22, 2025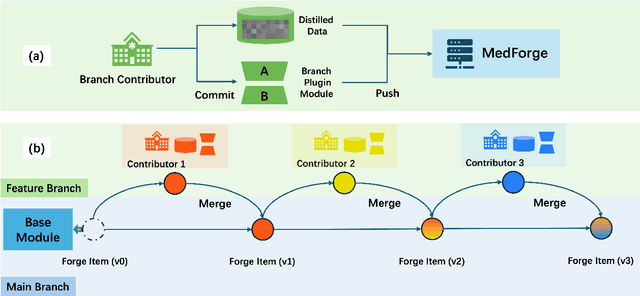
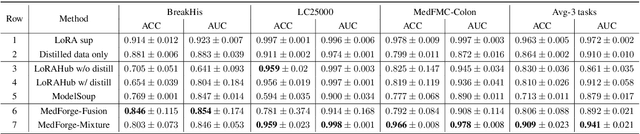
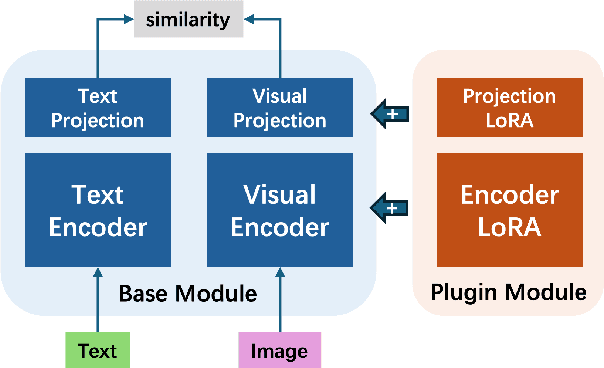
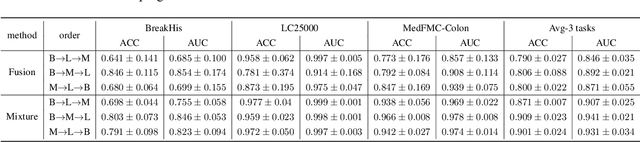
Abstract:Foundational models (FMs) have made significant strides in the healthcare domain. Yet the data silo challenge and privacy concern remain in healthcare systems, hindering safe medical data sharing and collaborative model development among institutions. The collection and curation of scalable clinical datasets increasingly become the bottleneck for training strong FMs. In this study, we propose Medical Foundation Models Merging (MedForge), a cooperative framework enabling a community-driven medical foundation model development, meanwhile preventing the information leakage of raw patient data and mitigating synchronization model development issues across clinical institutions. MedForge offers a bottom-up model construction mechanism by flexibly merging task-specific Low-Rank Adaptation (LoRA) modules, which can adapt to downstream tasks while retaining original model parameters. Through an asynchronous LoRA module integration scheme, the resulting composite model can progressively enhance its comprehensive performance on various clinical tasks. MedForge shows strong performance on multiple clinical datasets (e.g., breast cancer, lung cancer, and colon cancer) collected from different institutions. Our major findings highlight the value of collaborative foundation models in advancing multi-center clinical collaboration effectively and cohesively. Our code is publicly available at https://github.com/TanZheling/MedForge.
Improved Training Technique for Latent Consistency Models
Feb 03, 2025



Abstract:Consistency models are a new family of generative models capable of producing high-quality samples in either a single step or multiple steps. Recently, consistency models have demonstrated impressive performance, achieving results on par with diffusion models in the pixel space. However, the success of scaling consistency training to large-scale datasets, particularly for text-to-image and video generation tasks, is determined by performance in the latent space. In this work, we analyze the statistical differences between pixel and latent spaces, discovering that latent data often contains highly impulsive outliers, which significantly degrade the performance of iCT in the latent space. To address this, we replace Pseudo-Huber losses with Cauchy losses, effectively mitigating the impact of outliers. Additionally, we introduce a diffusion loss at early timesteps and employ optimal transport (OT) coupling to further enhance performance. Lastly, we introduce the adaptive scaling-$c$ scheduler to manage the robust training process and adopt Non-scaling LayerNorm in the architecture to better capture the statistics of the features and reduce outlier impact. With these strategies, we successfully train latent consistency models capable of high-quality sampling with one or two steps, significantly narrowing the performance gap between latent consistency and diffusion models. The implementation is released here: https://github.com/quandao10/sLCT/
RadAlign: Advancing Radiology Report Generation with Vision-Language Concept Alignment
Jan 13, 2025Abstract:Automated chest radiographs interpretation requires both accurate disease classification and detailed radiology report generation, presenting a significant challenge in the clinical workflow. Current approaches either focus on classification accuracy at the expense of interpretability or generate detailed but potentially unreliable reports through image captioning techniques. In this study, we present RadAlign, a novel framework that combines the predictive accuracy of vision-language models (VLMs) with the reasoning capabilities of large language models (LLMs). Inspired by the radiologist's workflow, RadAlign first employs a specialized VLM to align visual features with key medical concepts, achieving superior disease classification with an average AUC of 0.885 across multiple diseases. These recognized medical conditions, represented as text-based concepts in the aligned visual-language space, are then used to prompt LLM-based report generation. Enhanced by a retrieval-augmented generation mechanism that grounds outputs in similar historical cases, RadAlign delivers superior report quality with a GREEN score of 0.678, outperforming state-of-the-art methods' 0.634. Our framework maintains strong clinical interpretability while reducing hallucinations, advancing automated medical imaging and report analysis through integrated predictive and generative AI. Code is available at https://github.com/difeigu/RadAlign.
Rate-My-LoRA: Efficient and Adaptive Federated Model Tuning for Cardiac MRI Segmentation
Jan 06, 2025Abstract:Cardiovascular disease (CVD) and cardiac dyssynchrony are major public health problems in the United States. Precise cardiac image segmentation is crucial for extracting quantitative measures that help categorize cardiac dyssynchrony. However, achieving high accuracy often depends on centralizing large datasets from different hospitals, which can be challenging due to privacy concerns. To solve this problem, Federated Learning (FL) is proposed to enable decentralized model training on such data without exchanging sensitive information. However, bandwidth limitations and data heterogeneity remain as significant challenges in conventional FL algorithms. In this paper, we propose a novel efficient and adaptive federate learning method for cardiac segmentation that improves model performance while reducing the bandwidth requirement. Our method leverages the low-rank adaptation (LoRA) to regularize model weight update and reduce communication overhead. We also propose a \mymethod{} aggregation technique to address data heterogeneity among clients. This technique adaptively penalizes the aggregated weights from different clients by comparing the validation accuracy in each client, allowing better generalization performance and fast local adaptation. In-client and cross-client evaluations on public cardiac MR datasets demonstrate the superiority of our method over other LoRA-based federate learning approaches.
Self-Corrected Flow Distillation for Consistent One-Step and Few-Step Text-to-Image Generation
Dec 22, 2024



Abstract:Flow matching has emerged as a promising framework for training generative models, demonstrating impressive empirical performance while offering relative ease of training compared to diffusion-based models. However, this method still requires numerous function evaluations in the sampling process. To address these limitations, we introduce a self-corrected flow distillation method that effectively integrates consistency models and adversarial training within the flow-matching framework. This work is a pioneer in achieving consistent generation quality in both few-step and one-step sampling. Our extensive experiments validate the effectiveness of our method, yielding superior results both quantitatively and qualitatively on CelebA-HQ and zero-shot benchmarks on the COCO dataset. Our implementation is released at https://github.com/VinAIResearch/SCFlow
VerSe: Integrating Multiple Queries as Prompts for Versatile Cardiac MRI Segmentation
Dec 20, 2024Abstract:Despite the advances in learning-based image segmentation approach, the accurate segmentation of cardiac structures from magnetic resonance imaging (MRI) remains a critical challenge. While existing automatic segmentation methods have shown promise, they still require extensive manual corrections of the segmentation results by human experts, particularly in complex regions such as the basal and apical parts of the heart. Recent efforts have been made on developing interactive image segmentation methods that enable human-in-the-loop learning. However, they are semi-automatic and inefficient, due to their reliance on click-based prompts, especially for 3D cardiac MRI volumes. To address these limitations, we propose VerSe, a Versatile Segmentation framework to unify automatic and interactive segmentation through mutiple queries. Our key innovation lies in the joint learning of object and click queries as prompts for a shared segmentation backbone. VerSe supports both fully automatic segmentation, through object queries, and interactive mask refinement, by providing click queries when needed. With the proposed integrated prompting scheme, VerSe demonstrates significant improvement in performance and efficiency over existing methods, on both cardiac MRI and out-of-distribution medical imaging datasets. The code is available at https://github.com/bangwayne/Verse.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge