Leon Axel
Rate-My-LoRA: Efficient and Adaptive Federated Model Tuning for Cardiac MRI Segmentation
Jan 06, 2025Abstract:Cardiovascular disease (CVD) and cardiac dyssynchrony are major public health problems in the United States. Precise cardiac image segmentation is crucial for extracting quantitative measures that help categorize cardiac dyssynchrony. However, achieving high accuracy often depends on centralizing large datasets from different hospitals, which can be challenging due to privacy concerns. To solve this problem, Federated Learning (FL) is proposed to enable decentralized model training on such data without exchanging sensitive information. However, bandwidth limitations and data heterogeneity remain as significant challenges in conventional FL algorithms. In this paper, we propose a novel efficient and adaptive federate learning method for cardiac segmentation that improves model performance while reducing the bandwidth requirement. Our method leverages the low-rank adaptation (LoRA) to regularize model weight update and reduce communication overhead. We also propose a \mymethod{} aggregation technique to address data heterogeneity among clients. This technique adaptively penalizes the aggregated weights from different clients by comparing the validation accuracy in each client, allowing better generalization performance and fast local adaptation. In-client and cross-client evaluations on public cardiac MR datasets demonstrate the superiority of our method over other LoRA-based federate learning approaches.
VerSe: Integrating Multiple Queries as Prompts for Versatile Cardiac MRI Segmentation
Dec 20, 2024Abstract:Despite the advances in learning-based image segmentation approach, the accurate segmentation of cardiac structures from magnetic resonance imaging (MRI) remains a critical challenge. While existing automatic segmentation methods have shown promise, they still require extensive manual corrections of the segmentation results by human experts, particularly in complex regions such as the basal and apical parts of the heart. Recent efforts have been made on developing interactive image segmentation methods that enable human-in-the-loop learning. However, they are semi-automatic and inefficient, due to their reliance on click-based prompts, especially for 3D cardiac MRI volumes. To address these limitations, we propose VerSe, a Versatile Segmentation framework to unify automatic and interactive segmentation through mutiple queries. Our key innovation lies in the joint learning of object and click queries as prompts for a shared segmentation backbone. VerSe supports both fully automatic segmentation, through object queries, and interactive mask refinement, by providing click queries when needed. With the proposed integrated prompting scheme, VerSe demonstrates significant improvement in performance and efficiency over existing methods, on both cardiac MRI and out-of-distribution medical imaging datasets. The code is available at https://github.com/bangwayne/Verse.
Learning Volumetric Neural Deformable Models to Recover 3D Regional Heart Wall Motion from Multi-Planar Tagged MRI
Nov 21, 2024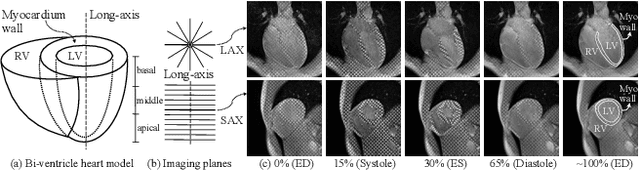
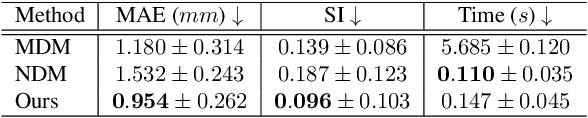
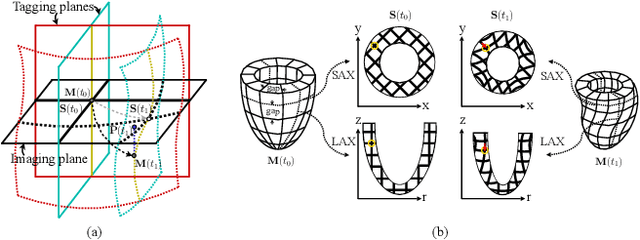
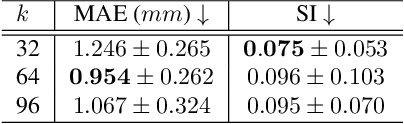
Abstract:Multi-planar tagged MRI is the gold standard for regional heart wall motion evaluation. However, accurate recovery of the 3D true heart wall motion from a set of 2D apparent motion cues is challenging, due to incomplete sampling of the true motion and difficulty in information fusion from apparent motion cues observed on multiple imaging planes. To solve these challenges, we introduce a novel class of volumetric neural deformable models ($\upsilon$NDMs). Our $\upsilon$NDMs represent heart wall geometry and motion through a set of low-dimensional global deformation parameter functions and a diffeomorphic point flow regularized local deformation field. To learn such global and local deformation for 2D apparent motion mapping to 3D true motion, we design a hybrid point transformer, which incorporates both point cross-attention and self-attention mechanisms. While use of point cross-attention can learn to fuse 2D apparent motion cues into material point true motion hints, point self-attention hierarchically organised as an encoder-decoder structure can further learn to refine these hints and map them into 3D true motion. We have performed experiments on a large cohort of synthetic 3D regional heart wall motion dataset. The results demonstrated the high accuracy of our method for the recovery of dense 3D true motion from sparse 2D apparent motion cues. Project page is at https://github.com/DeepTag/VolumetricNeuralDeformableModels.
Continuous Spatio-Temporal Memory Networks for 4D Cardiac Cine MRI Segmentation
Oct 30, 2024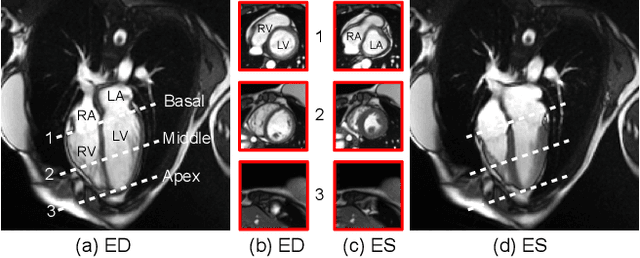
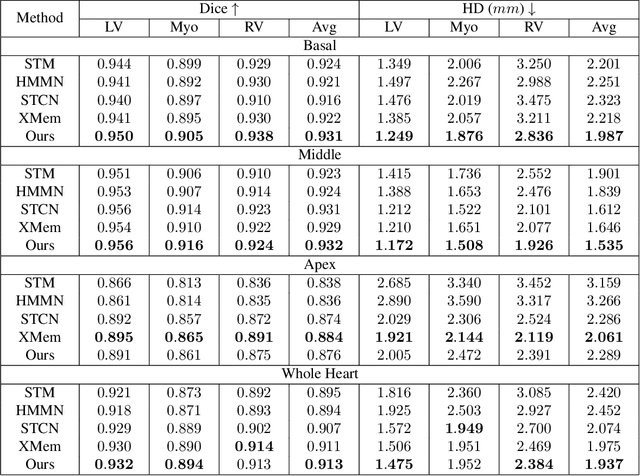
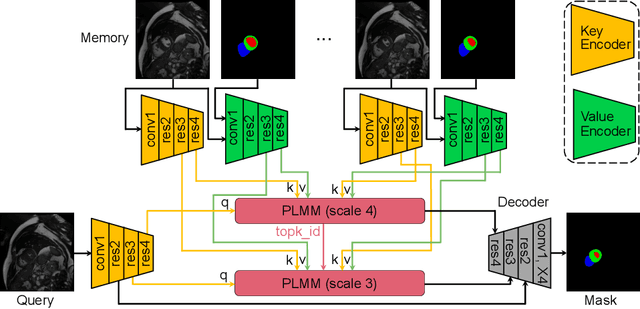
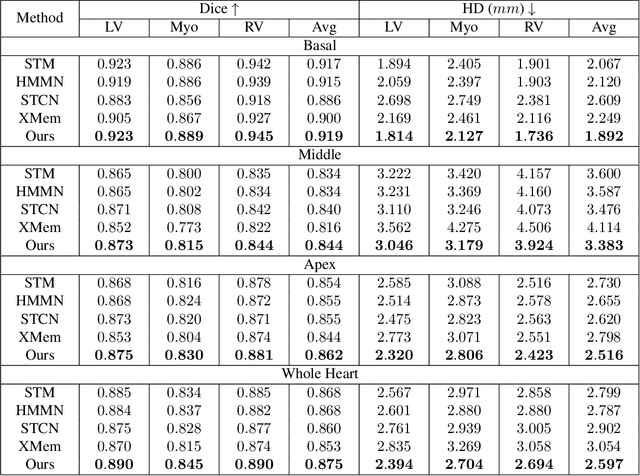
Abstract:Current cardiac cine magnetic resonance image (cMR) studies focus on the end diastole (ED) and end systole (ES) phases, while ignoring the abundant temporal information in the whole image sequence. This is because whole sequence segmentation is currently a tedious process and inaccurate. Conventional whole sequence segmentation approaches first estimate the motion field between frames, which is then used to propagate the mask along the temporal axis. However, the mask propagation results could be prone to error, especially for the basal and apex slices, where through-plane motion leads to significant morphology and structural change during the cardiac cycle. Inspired by recent advances in video object segmentation (VOS), based on spatio-temporal memory (STM) networks, we propose a continuous STM (CSTM) network for semi-supervised whole heart and whole sequence cMR segmentation. Our CSTM network takes full advantage of the spatial, scale, temporal and through-plane continuity prior of the underlying heart anatomy structures, to achieve accurate and fast 4D segmentation. Results of extensive experiments across multiple cMR datasets show that our method can improve the 4D cMR segmentation performance, especially for the hard-to-segment regions.
Fill the K-Space and Refine the Image: Prompting for Dynamic and Multi-Contrast MRI Reconstruction
Sep 25, 2023



Abstract:The key to dynamic or multi-contrast magnetic resonance imaging (MRI) reconstruction lies in exploring inter-frame or inter-contrast information. Currently, the unrolled model, an approach combining iterative MRI reconstruction steps with learnable neural network layers, stands as the best-performing method for MRI reconstruction. However, there are two main limitations to overcome: firstly, the unrolled model structure and GPU memory constraints restrict the capacity of each denoising block in the network, impeding the effective extraction of detailed features for reconstruction; secondly, the existing model lacks the flexibility to adapt to variations in the input, such as different contrasts, resolutions or views, necessitating the training of separate models for each input type, which is inefficient and may lead to insufficient reconstruction. In this paper, we propose a two-stage MRI reconstruction pipeline to address these limitations. The first stage involves filling the missing k-space data, which we approach as a physics-based reconstruction problem. We first propose a simple yet efficient baseline model, which utilizes adjacent frames/contrasts and channel attention to capture the inherent inter-frame/-contrast correlation. Then, we extend the baseline model to a prompt-based learning approach, PromptMR, for all-in-one MRI reconstruction from different views, contrasts, adjacent types, and acceleration factors. The second stage is to refine the reconstruction from the first stage, which we treat as a general video restoration problem to further fuse features from neighboring frames/contrasts in the image domain. Extensive experiments show that our proposed method significantly outperforms previous state-of-the-art accelerated MRI reconstruction methods.
DMCVR: Morphology-Guided Diffusion Model for 3D Cardiac Volume Reconstruction
Aug 18, 2023Abstract:Accurate 3D cardiac reconstruction from cine magnetic resonance imaging (cMRI) is crucial for improved cardiovascular disease diagnosis and understanding of the heart's motion. However, current cardiac MRI-based reconstruction technology used in clinical settings is 2D with limited through-plane resolution, resulting in low-quality reconstructed cardiac volumes. To better reconstruct 3D cardiac volumes from sparse 2D image stacks, we propose a morphology-guided diffusion model for 3D cardiac volume reconstruction, DMCVR, that synthesizes high-resolution 2D images and corresponding 3D reconstructed volumes. Our method outperforms previous approaches by conditioning the cardiac morphology on the generative model, eliminating the time-consuming iterative optimization process of the latent code, and improving generation quality. The learned latent spaces provide global semantics, local cardiac morphology and details of each 2D cMRI slice with highly interpretable value to reconstruct 3D cardiac shape. Our experiments show that DMCVR is highly effective in several aspects, such as 2D generation and 3D reconstruction performance. With DMCVR, we can produce high-resolution 3D cardiac MRI reconstructions, surpassing current techniques. Our proposed framework has great potential for improving the accuracy of cardiac disease diagnosis and treatment planning. Code can be accessed at https://github.com/hexiaoxiao-cs/DMCVR.
Neural Deformable Models for 3D Bi-Ventricular Heart Shape Reconstruction and Modeling from 2D Sparse Cardiac Magnetic Resonance Imaging
Jul 15, 2023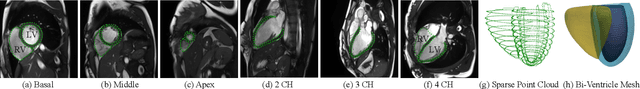
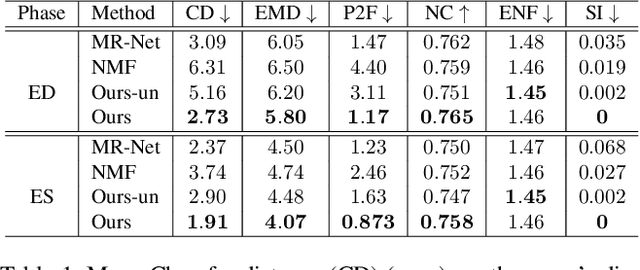
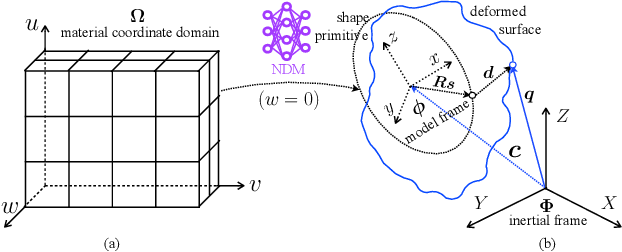

Abstract:We propose a novel neural deformable model (NDM) targeting at the reconstruction and modeling of 3D bi-ventricular shape of the heart from 2D sparse cardiac magnetic resonance (CMR) imaging data. We model the bi-ventricular shape using blended deformable superquadrics, which are parameterized by a set of geometric parameter functions and are capable of deforming globally and locally. While global geometric parameter functions and deformations capture gross shape features from visual data, local deformations, parameterized as neural diffeomorphic point flows, can be learned to recover the detailed heart shape.Different from iterative optimization methods used in conventional deformable model formulations, NDMs can be trained to learn such geometric parameter functions, global and local deformations from a shape distribution manifold. Our NDM can learn to densify a sparse cardiac point cloud with arbitrary scales and generate high-quality triangular meshes automatically. It also enables the implicit learning of dense correspondences among different heart shape instances for accurate cardiac shape registration. Furthermore, the parameters of NDM are intuitive, and can be used by a physician without sophisticated post-processing. Experimental results on a large CMR dataset demonstrate the improved performance of NDM over conventional methods.
DeepRecon: Joint 2D Cardiac Segmentation and 3D Volume Reconstruction via A Structure-Specific Generative Method
Jun 14, 2022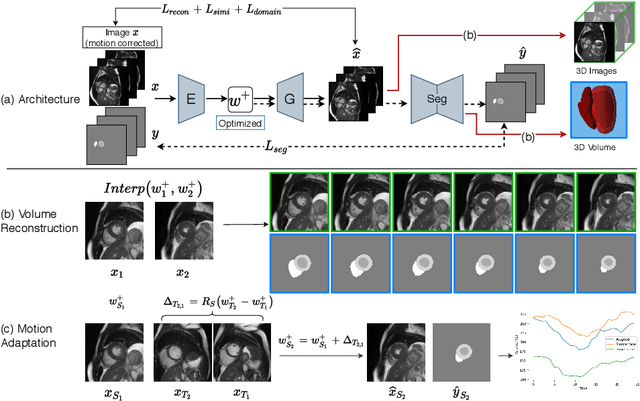
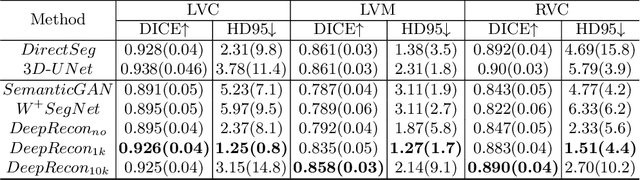
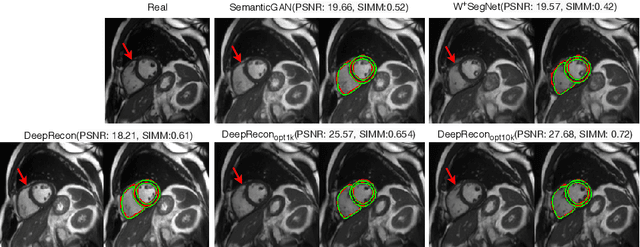
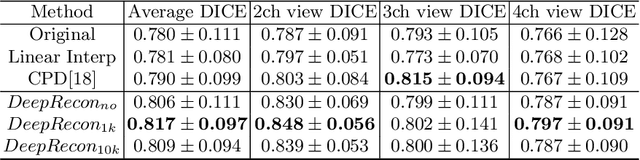
Abstract:Joint 2D cardiac segmentation and 3D volume reconstruction are fundamental to building statistical cardiac anatomy models and understanding functional mechanisms from motion patterns. However, due to the low through-plane resolution of cine MR and high inter-subject variance, accurately segmenting cardiac images and reconstructing the 3D volume are challenging. In this study, we propose an end-to-end latent-space-based framework, DeepRecon, that generates multiple clinically essential outcomes, including accurate image segmentation, synthetic high-resolution 3D image, and 3D reconstructed volume. Our method identifies the optimal latent representation of the cine image that contains accurate semantic information for cardiac structures. In particular, our model jointly generates synthetic images with accurate semantic information and segmentation of the cardiac structures using the optimal latent representation. We further explore downstream applications of 3D shape reconstruction and 4D motion pattern adaptation by the different latent-space manipulation strategies.The simultaneously generated high-resolution images present a high interpretable value to assess the cardiac shape and motion.Experimental results demonstrate the effectiveness of our approach on multiple fronts including 2D segmentation, 3D reconstruction, downstream 4D motion pattern adaption performance.
Learned Half-Quadratic Splitting Network for Magnetic Resonance Image Reconstruction
Dec 21, 2021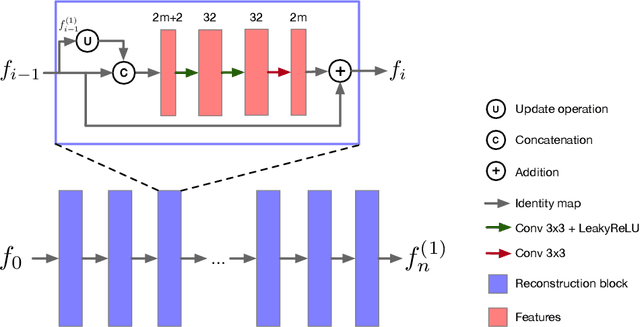
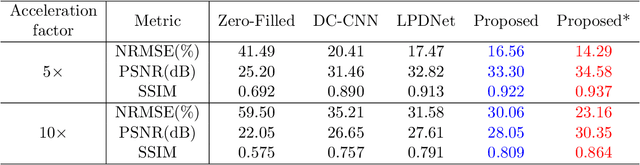
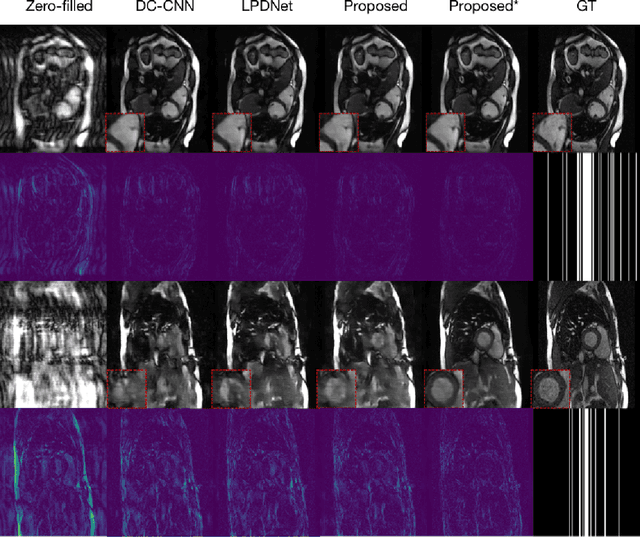

Abstract:Magnetic Resonance (MR) image reconstruction from highly undersampled $k$-space data is critical in accelerated MR imaging (MRI) techniques. In recent years, deep learning-based methods have shown great potential in this task. This paper proposes a learned half-quadratic splitting algorithm for MR image reconstruction and implements the algorithm in an unrolled deep learning network architecture. We compare the performance of our proposed method on a public cardiac MR dataset against DC-CNN and LPDNet, and our method outperforms other methods in both quantitative results and qualitative results with fewer model parameters and faster reconstruction speed. Finally, we enlarge our model to achieve superior reconstruction quality, and the improvement is $1.76$ dB and $2.74$ dB over LPDNet in peak signal-to-noise ratio on $5\times$ and $10\times$ acceleration, respectively. Code for our method is publicly available at https://github.com/hellopipu/HQS-Net.
DeepTag: An Unsupervised Deep Learning Method for Motion Tracking on Cardiac Tagging Magnetic Resonance Images
Mar 29, 2021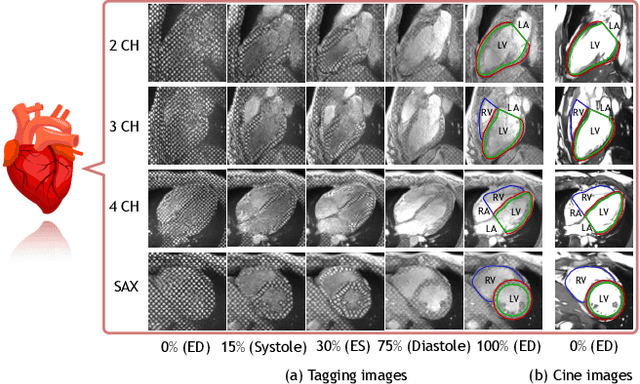
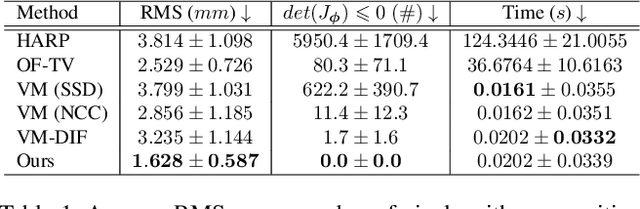
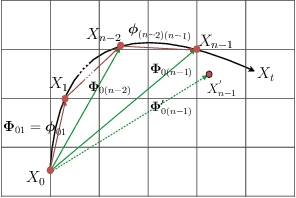
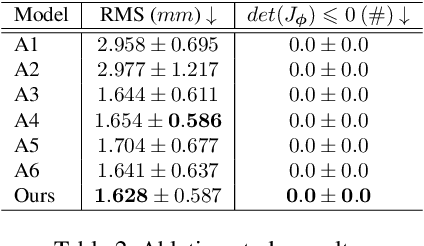
Abstract:Cardiac tagging magnetic resonance imaging (t-MRI) is the gold standard for regional myocardium deformation and cardiac strain estimation. However, this technique has not been widely used in clinical diagnosis, as a result of the difficulty of motion tracking encountered with t-MRI images. In this paper, we propose a novel deep learning-based fully unsupervised method for in vivo motion tracking on t-MRI images. We first estimate the motion field (INF) between any two consecutive t-MRI frames by a bi-directional generative diffeomorphic registration neural network. Using this result, we then estimate the Lagrangian motion field between the reference frame and any other frame through a differentiable composition layer. By utilizing temporal information to perform reasonable estimations on spatio-temporal motion fields, this novel method provides a useful solution for motion tracking and image registration in dynamic medical imaging. Our method has been validated on a representative clinical t-MRI dataset; the experimental results show that our method is superior to conventional motion tracking methods in terms of landmark tracking accuracy and inference efficiency.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge