Mikael Kanski
Neural Deformable Models for 3D Bi-Ventricular Heart Shape Reconstruction and Modeling from 2D Sparse Cardiac Magnetic Resonance Imaging
Jul 15, 2023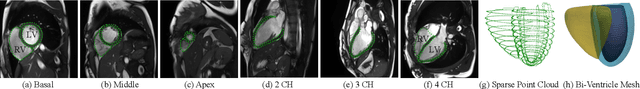
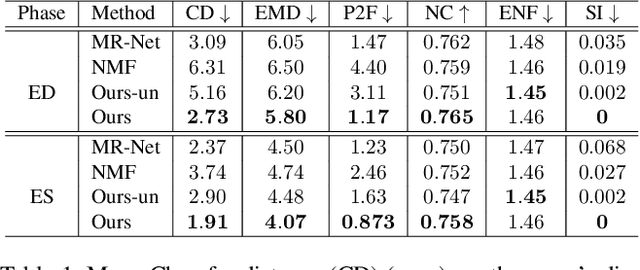
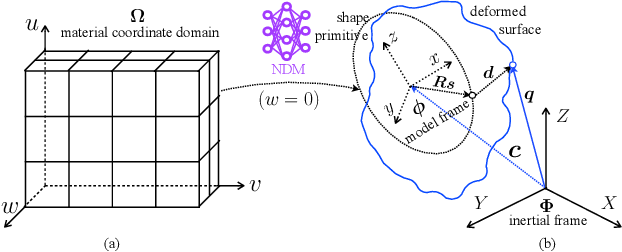

Abstract:We propose a novel neural deformable model (NDM) targeting at the reconstruction and modeling of 3D bi-ventricular shape of the heart from 2D sparse cardiac magnetic resonance (CMR) imaging data. We model the bi-ventricular shape using blended deformable superquadrics, which are parameterized by a set of geometric parameter functions and are capable of deforming globally and locally. While global geometric parameter functions and deformations capture gross shape features from visual data, local deformations, parameterized as neural diffeomorphic point flows, can be learned to recover the detailed heart shape.Different from iterative optimization methods used in conventional deformable model formulations, NDMs can be trained to learn such geometric parameter functions, global and local deformations from a shape distribution manifold. Our NDM can learn to densify a sparse cardiac point cloud with arbitrary scales and generate high-quality triangular meshes automatically. It also enables the implicit learning of dense correspondences among different heart shape instances for accurate cardiac shape registration. Furthermore, the parameters of NDM are intuitive, and can be used by a physician without sophisticated post-processing. Experimental results on a large CMR dataset demonstrate the improved performance of NDM over conventional methods.
DeepRecon: Joint 2D Cardiac Segmentation and 3D Volume Reconstruction via A Structure-Specific Generative Method
Jun 14, 2022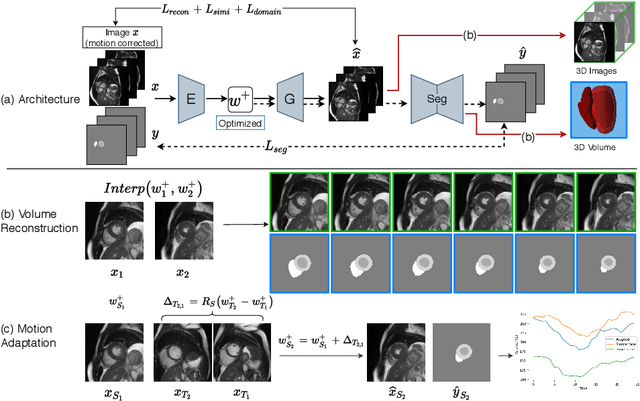
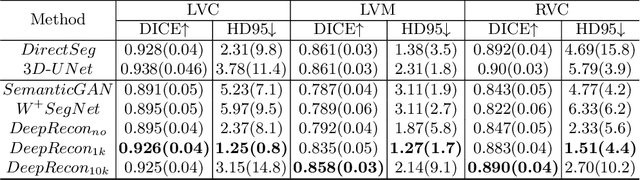
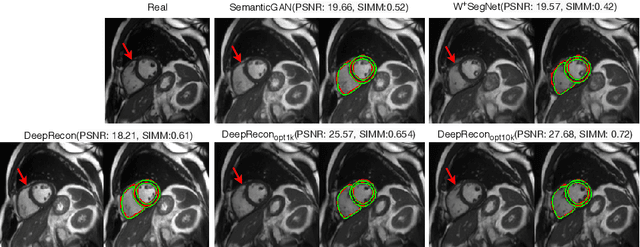
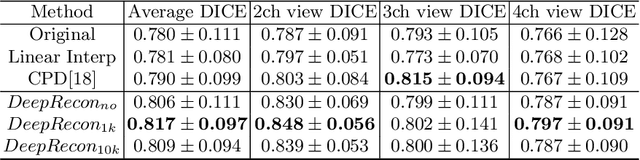
Abstract:Joint 2D cardiac segmentation and 3D volume reconstruction are fundamental to building statistical cardiac anatomy models and understanding functional mechanisms from motion patterns. However, due to the low through-plane resolution of cine MR and high inter-subject variance, accurately segmenting cardiac images and reconstructing the 3D volume are challenging. In this study, we propose an end-to-end latent-space-based framework, DeepRecon, that generates multiple clinically essential outcomes, including accurate image segmentation, synthetic high-resolution 3D image, and 3D reconstructed volume. Our method identifies the optimal latent representation of the cine image that contains accurate semantic information for cardiac structures. In particular, our model jointly generates synthetic images with accurate semantic information and segmentation of the cardiac structures using the optimal latent representation. We further explore downstream applications of 3D shape reconstruction and 4D motion pattern adaptation by the different latent-space manipulation strategies.The simultaneously generated high-resolution images present a high interpretable value to assess the cardiac shape and motion.Experimental results demonstrate the effectiveness of our approach on multiple fronts including 2D segmentation, 3D reconstruction, downstream 4D motion pattern adaption performance.
DeepTag: An Unsupervised Deep Learning Method for Motion Tracking on Cardiac Tagging Magnetic Resonance Images
Mar 29, 2021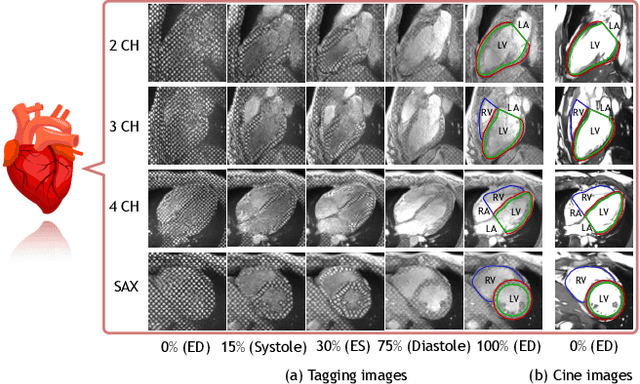
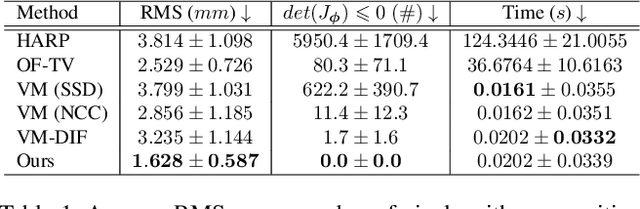
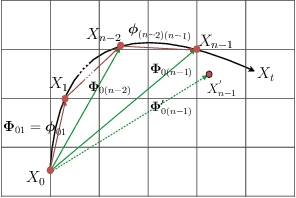
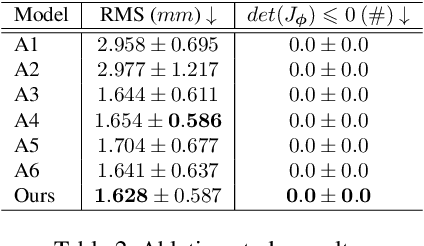
Abstract:Cardiac tagging magnetic resonance imaging (t-MRI) is the gold standard for regional myocardium deformation and cardiac strain estimation. However, this technique has not been widely used in clinical diagnosis, as a result of the difficulty of motion tracking encountered with t-MRI images. In this paper, we propose a novel deep learning-based fully unsupervised method for in vivo motion tracking on t-MRI images. We first estimate the motion field (INF) between any two consecutive t-MRI frames by a bi-directional generative diffeomorphic registration neural network. Using this result, we then estimate the Lagrangian motion field between the reference frame and any other frame through a differentiable composition layer. By utilizing temporal information to perform reasonable estimations on spatio-temporal motion fields, this novel method provides a useful solution for motion tracking and image registration in dynamic medical imaging. Our method has been validated on a representative clinical t-MRI dataset; the experimental results show that our method is superior to conventional motion tracking methods in terms of landmark tracking accuracy and inference efficiency.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge